
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 1,982,191 – 1,982,293 |
| Length | 102 |
| Max. P | 0.872608 |

| Location | 1,982,191 – 1,982,293 |
|---|---|
| Length | 102 |
| Sequences | 12 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 80.31 |
| Shannon entropy | 0.41100 |
| G+C content | 0.38592 |
| Mean single sequence MFE | -21.02 |
| Consensus MFE | -16.58 |
| Energy contribution | -16.75 |
| Covariance contribution | 0.17 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.24 |
| Structure conservation index | 0.79 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.01 |
| SVM RNA-class probability | 0.872608 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
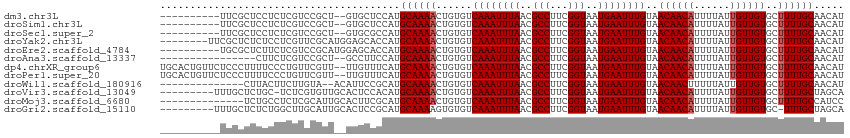
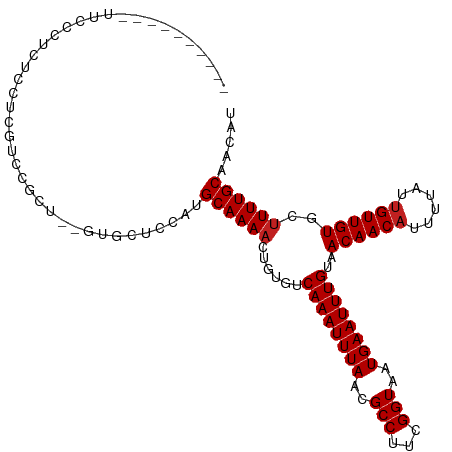
>dm3.chr3L 1982191 102 - 24543557 ----------UUCGCUCCUCUCGUCCGCU--GUGCUCCAUGCAAAACUGUGUCAAAUUUAACGCCUUCGGUAAUGAAUUUGUAACAACAUUUUAUUGUUGUGCUUUUGCAACAU ----------...................--........(((((((......((((((((..(((...)))..))))))))..((((((......))))))..))))))).... ( -18.30, z-score = -0.81, R) >droSim1.chr3L 1551380 102 - 22553184 ----------UUCGCUCCUCUCGUCCGCU--GUGCUCCAUGCAAAACUGUGUCAAAUUUAACGCCUUCGGUAAUGAAUUUGUAACAACAUUUUAUUGUUGUGCUUUUGCAACAU ----------...................--........(((((((......((((((((..(((...)))..))))))))..((((((......))))))..))))))).... ( -18.30, z-score = -0.81, R) >droSec1.super_2 2007386 102 - 7591821 ----------UUCGCUCCUCUCGUCCGCU--GUGCGCCAUGCAAAACUGUGUCAAAUUUAACGCCUUCGGUAAUGAAUUUGUAACAACAUUUUAUUGUUGUGCUUUUGCAACAU ----------..(((.(..(......)..--).)))...(((((((......((((((((..(((...)))..))))))))..((((((......))))))..))))))).... ( -19.70, z-score = -0.55, R) >droYak2.chr3L 1936816 106 - 24197627 --------UUCGCUCUCUCCUCGUUCGCAUGGAGCACCAUGCAAAACUGUGUCAAAUUUAACGCCUUCGGUAAUGAAUUUGUAACAACAUUUUAUUGUUGUGCUUUUGCAACAU --------..............(((.(((.(((((((...((((....((((((((((((..(((...)))..)))))))).....))))....)))).))))))))))))).. ( -23.80, z-score = -1.63, R) >droEre2.scaffold_4784 1971159 104 - 25762168 ----------UGCGCUCUUCUCGUCCGCAUGGAGCACCAUGCAAAACUGUGUCAAAUUUAACGCCUUCGGUAAUGAAUUUGUAACAACAUUUUAUUGUUGUGCUUUUGCAACAU ----------...(((((...(....)...)))))....(((((((......((((((((..(((...)))..))))))))..((((((......))))))..))))))).... ( -24.70, z-score = -1.21, R) >droAna3.scaffold_13337 22441662 96 + 23293914 ----------------CUUCUCGUCCGCU--GCCUUCCAUGCAAAACUGUGUCAAAUUUAACGCCUUCGGUAAUGAAUUUGUAACAACAUUUUAUUGUUGUGCUUUUGCAACAU ----------------.............--........(((((((......((((((((..(((...)))..))))))))..((((((......))))))..))))))).... ( -18.30, z-score = -1.50, R) >dp4.chrXR_group6 11857601 112 - 13314419 UGCACUGUUCUCCCUUUUCCCUGUUCGUU--UUGUUUCAUGCAAAACUGUGUCAAAUUUAACGCCUUCGGUAAUGAAUUUGUAACAACAUUUUAUUGUUGUGCUUUUGCAACAU ((((......................(((--((((.....))))))).....((((((((..(((...)))..))))))))..((((((......)))))).....)))).... ( -21.20, z-score = -1.72, R) >droPer1.super_20 1781357 112 - 1872136 UGCACUGUUCUCCCUUUUCCCUGUUCGUU--UUGUUUCAUGCAAAACUGUGUCAAAUUUAACGCCUUCGGUAAUGAAUUUGUAACAACAUUUUAUUGUUGUGCUUUUGCAACAU ((((......................(((--((((.....))))))).....((((((((..(((...)))..))))))))..((((((......)))))).....)))).... ( -21.20, z-score = -1.72, R) >droWil1.scaffold_180916 1250477 98 + 2700594 --------------CUUACUUCUUGUA--ACAUUCCGCAUGCAAAACUGUGUCAAAUUUAACGCCUUCGGUAAUGAAUUUGUAACAACUUUUUAUUGUUGUGCUUUUGCAACAU --------------.((((.....)))--).........(((((((......((((((((..(((...)))..))))))))..(((((........)))))..))))))).... ( -17.80, z-score = -1.30, R) >droVir3.scaffold_13049 13080729 104 + 25233164 ---------UUUGCUCUGC-UCUCGUGUUGCACUCCACAUGCAAAACUGUGUCAAAUUUAACGCCUUCGGUAAUGAAUUUGUAACAACAUUUUAUUGUUGUGCUUUUGCUAGCA ---------..((((..((-....((.(((((.......))))).)).....((((((((..(((...)))..))))))))..((((((......))))))......)).)))) ( -21.40, z-score = -0.67, R) >droMoj3.scaffold_6680 8691312 100 - 24764193 --------------UCUGCCUCUCGCAUUGCACUUCGCAUGCAAAACUGUGUCAAAUUUAACGCCUUCGGUAAUGAAUUUGUAACAACAUUUUAUUGUUGUGCUUUUGCCAUCC --------------..(((.....))).(((.....))).((((((......((((((((..(((...)))..))))))))..((((((......))))))..))))))..... ( -19.10, z-score = -1.01, R) >droGri2.scaffold_15110 12997294 104 + 24565398 ---------UUUGCUCUCUGGCUUGCAUUGCACUCCGCAUGCAAAAGUGUGUCAAAUUUAACGCCUUCGGUAAUGAAUUUGUAACAACAUUUUAUUGUUGUGC-UUUGCUAGCA ---------...........(((.(((..((((...(((....(((((((..((((((((..(((...)))..)))))))).....)))))))..))).))))-..))).))). ( -28.40, z-score = -1.99, R) >consensus __________UUCCCUCUCCUCGUCCGCU__GUGCUCCAUGCAAAACUGUGUCAAAUUUAACGCCUUCGGUAAUGAAUUUGUAACAACAUUUUAUUGUUGUGCUUUUGCAACAU ........................................((((((......((((((((..(((...)))..))))))))..((((((......))))))..))))))..... (-16.58 = -16.75 + 0.17)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:56:51 2011