| Sequence ID | dm3.chr3L |
|---|---|
| Location | 1,928,298 – 1,928,392 |
| Length | 94 |
| Max. P | 0.979321 |

| Location | 1,928,298 – 1,928,392 |
|---|---|
| Length | 94 |
| Sequences | 12 |
| Columns | 121 |
| Reading direction | forward |
| Mean pairwise identity | 81.13 |
| Shannon entropy | 0.36651 |
| G+C content | 0.42104 |
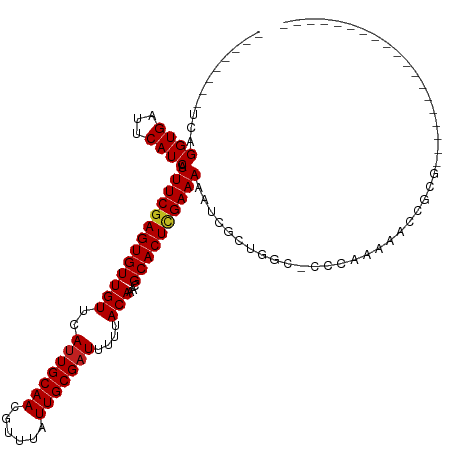
| Mean single sequence MFE | -27.07 |
| Consensus MFE | -20.05 |
| Energy contribution | -20.15 |
| Covariance contribution | 0.10 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.18 |
| Structure conservation index | 0.74 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.02 |
| SVM RNA-class probability | 0.979321 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
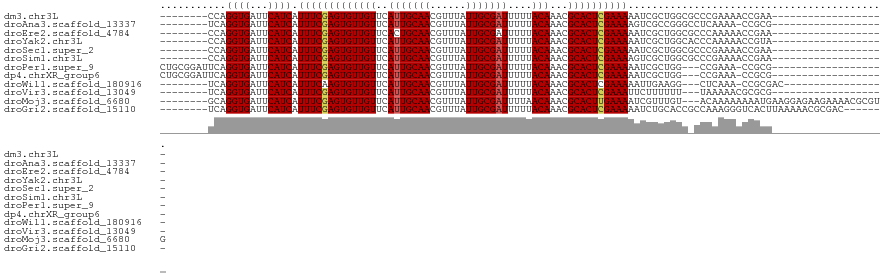
>dm3.chr3L 1928298 94 + 24543557 --------CCAGGUGAUUCAUCAUUUCGAGUGUUGUUCAUUGCAACGUUUAUUGCGAUUUUUACAAACGCACUCGAAAAAUCGCUGGCGCCCGAAAACCGAA------------------- --------((((.(((((.....(((((((((((((..(((((((......)))))))....)))...)))))))))))))))))))...............------------------- ( -26.60, z-score = -1.97, R) >droAna3.scaffold_13337 22390000 93 - 23293914 --------UCAGGUGAUUCAUCAUUUCGAGUGUUGUUCAUUGCAACGUUUAUUGCGAUUUUUACAAACGCACUCGAAAAGUCGCCGGGCCUCAAAA-CCGCG------------------- --------...(((((((.....(((((((((((((..(((((((......)))))))....)))...)))))))))))))))))((.........-))...------------------- ( -29.00, z-score = -3.00, R) >droEre2.scaffold_4784 1917276 94 + 25762168 --------CCAGGUGAUUCAUCAUUUCGAGUGUUGUUCACUGCAACGUUUAUUGCGAUUUUUACAAACGCACUCGAAAAAUCGCUGGCGCCCAAAAACCGAA------------------- --------((((.(((((.....(((((((((((((....(((((......)))))......)))...)))))))))))))))))))...............------------------- ( -23.70, z-score = -1.44, R) >droYak2.chr3L 1882491 94 + 24197627 --------CCAGGUGAUUCAUCAUUUCGAGUGUUGUUCAUUGCAACGUUUAUUGCGAUUUUUACAAACGCACUCGAAAAAUCGCUGGCACCCAAAAACCGUA------------------- --------((((.(((((.....(((((((((((((..(((((((......)))))))....)))...)))))))))))))))))))...............------------------- ( -26.60, z-score = -2.91, R) >droSec1.super_2 1953726 94 + 7591821 --------CCAGGUGAUUCAUCAUUUCGAGUGUUGUUCAUUGCAACGUUUAUUGCGAUUUUUACAAACGCACUCGAAAAAUCGCUGGCGCCCGAAAACCGAA------------------- --------((((.(((((.....(((((((((((((..(((((((......)))))))....)))...)))))))))))))))))))...............------------------- ( -26.60, z-score = -1.97, R) >droSim1.chr3L 1483418 94 + 22553184 --------CCAGGUGAUUCAUCAUUUCGAGUGUUGUUCAUUGCAACGUUUAUUGCGAUUUUUACAAACGCACUCGAAAAGUCGCUGGCGCCCGAAAACCGAA------------------- --------((((.(((((.....(((((((((((((..(((((((......)))))))....)))...)))))))))))))))))))...............------------------- ( -26.60, z-score = -1.72, R) >droPer1.super_9 3423325 98 + 3637205 CUGCGGAUUCAGGUGAUUCAUCAUUUCGAGUGUUGUUCAUUGCAACGUUUAUUGCGAUUUUUACAAACGCACUCGAAAAAUCGCUGG---CCGAAA-CCGCG------------------- ..((((.(((.(((((((.....(((((((((((((..(((((((......)))))))....)))...)))))))))))))))))..---..))).-)))).------------------- ( -34.70, z-score = -3.70, R) >dp4.chrXR_group6 5114184 98 + 13314419 CUGCGGAUUCAGGUGAUUCAUCAUUUCGAGUGUUGUUCAUUGCAACGUUUAUUGCGAUUUUUACAAACGCACUCGAAAAAUCGCUGG---CCGAAA-CCGCG------------------- ..((((.(((.(((((((.....(((((((((((((..(((((((......)))))))....)))...)))))))))))))))))..---..))).-)))).------------------- ( -34.70, z-score = -3.70, R) >droWil1.scaffold_180916 1177301 92 - 2700594 --------UCAGGUGAUUCAUCAUUUCAAGUGUUGUUCAUUGCAACGUUUAUUGCGAUUUUUACAAACGCACUCGAAAAAUUGAAGG---CUCAAA-CCGCGAC----------------- --------...(((..((((...((((.((((((((..(((((((......)))))))....)))...))))).))))...))))..---.....)-)).....----------------- ( -19.30, z-score = -0.49, R) >droVir3.scaffold_13049 15856420 91 - 25233164 --------UCAGGUGAUUCAUCAUUUCGAGUGUUGUUCAUUGCAACGUUUAUUGCGAUUUUUACAAACGCACUCGAAAUUCUUUUUU---UAAAAACGCGCG------------------- --------...((((...))))((((((((((((((..(((((((......)))))))....)))...)))))))))))........---............------------------- ( -21.40, z-score = -1.77, R) >droMoj3.scaffold_6680 11378832 110 + 24764193 --------GCAGGUGAUUCAUCAUUUCGAGUGUUGUUCAUUGCAACGUUUAUUGCGAUUUUAACAAACGCACUUGAAAAUCGUUUGU---ACAAAAAAAAUGAAGGAGAAGAAAACGCGUG --------.((.(((.(((.((.((((((((((((((.(((((((......)))))))...))))...)))))))))).((((((..---.......))))))....)).)))..))).)) ( -26.70, z-score = -1.08, R) >droGri2.scaffold_15110 6784846 106 + 24565398 --------UCAGGUGAUUCAUCAUUUCGAGUGUUGUUCAUUGCAACGUUUAUUGCGAUUUUUACAAACGCACUCGAAAAAUCUGCACCGCCAAAGGGUCACUUAAAAACGCGAC------- --------..(((((((((....(((((((((((((..(((((((......)))))))....)))...)))))))))).....((...))....)))))))))...........------- ( -28.90, z-score = -2.44, R) >consensus ________UCAGGUGAUUCAUCAUUUCGAGUGUUGUUCAUUGCAACGUUUAUUGCGAUUUUUACAAACGCACUCGAAAAAUCGCUGGC_CCCAAAAACCGCG___________________ ...........((((...)))).(((((((((((((..(((((((......)))))))....)))...))))))))))........................................... (-20.05 = -20.15 + 0.10)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:56:49 2011