| Sequence ID | dm3.chr3L |
|---|---|
| Location | 1,925,639 – 1,925,741 |
| Length | 102 |
| Max. P | 0.961095 |

| Location | 1,925,639 – 1,925,741 |
|---|---|
| Length | 102 |
| Sequences | 12 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 67.79 |
| Shannon entropy | 0.68227 |
| G+C content | 0.48755 |
| Mean single sequence MFE | -29.83 |
| Consensus MFE | -19.50 |
| Energy contribution | -18.67 |
| Covariance contribution | -0.84 |
| Combinations/Pair | 1.67 |
| Mean z-score | -0.57 |
| Structure conservation index | 0.65 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.39 |
| SVM RNA-class probability | 0.935162 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
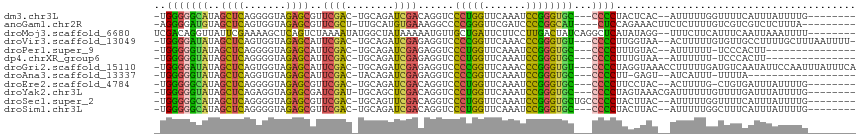
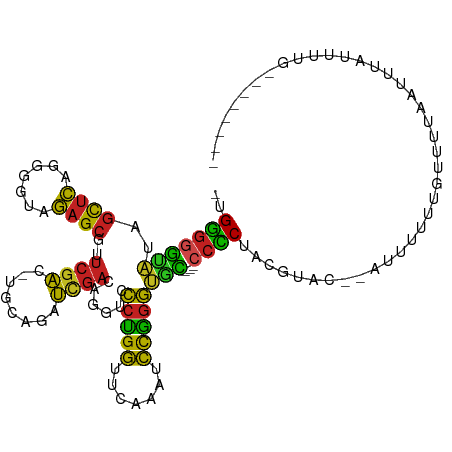
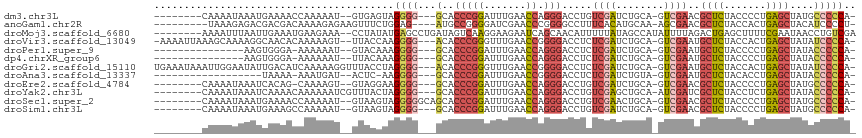
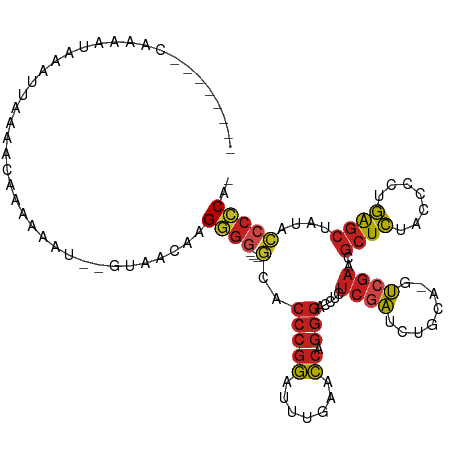
>dm3.chr3L 1925639 102 + 24543557 -UGGGGGCAUAGCUCAGGGGUAGAGCGUUCGAC-UGCAGAUCGACAGGUCCCUGGUUCAAAUCCGGGUGC---CCCCUACUCAC--AUUUUUGGUUUUCAUUUAUUUUG-------- -.(((((((..((((.......)))).......-....((((....))))(((((.......))))))))---)))).......--.......................-------- ( -30.10, z-score = -0.35, R) >anoGam1.chr2R 51280140 102 - 62725911 -AGGGGAUGUAGCUCAGUGGUAGAGCGUUCGCU-UUGCAUGUGAAAGGCCCCGGGUUCGAUCCCCGGCAU----CUCCAGAAACUUCUCUUUUGUCGUCGUCUCUUUA--------- -.((((((..(((((....(((((((....)))-))))........(....))))))..))))))(((..----(..(((((.......)))))..)..)))......--------- ( -30.00, z-score = -0.21, R) >droMoj3.scaffold_6680 11374934 107 + 24764193 UCGACAGGUUAUUCGAAAAGCUCAGUCUAAAAUAUGGCUAUAAAAAUGUUGCUGAUUCUUCCUUGACUAUCAGGCUCAUAUAGG--UUUCUUCAUUUCAAUUAAAUUUU-------- ..............((((((((.((((........))))......(((...(((((((......))..)))))...)))...))--))).)))................-------- ( -12.10, z-score = 1.45, R) >droVir3.scaffold_13049 145078 109 + 25233164 -UGGGGAUAUAGCUCAGUGGUAGAGCAUUCGAC-UGCAGAUCGAGAGGUCCCCGGUUCAAACCCGGGUGU---CCCCUUGGUAA--ACUUUUUGUGUUGCCUUUUGCUUUAAUUUU- -.(((((((..((((.......)))).......-....((((....))))(((((.......))))))))---))))..(((((--((.....)).)))))...............- ( -34.40, z-score = -1.34, R) >droPer1.super_9 3419204 94 + 3637205 -UGGGGGUAUAGCUCAGGGGUAGAGCAUUCGAC-UGCAGAUCGAGAGGUCCCUGGUUCAAAUCCGGGUGC---CCCCUUUGUAC--AUUUUUU-UCCCACUU--------------- -(((((((((((....((((((..(((......-))).((((....))))(((((.......))))))))---)))..))))))--.......-)))))...--------------- ( -30.61, z-score = -0.45, R) >dp4.chrXR_group6 5109246 94 + 13314419 -UGGGGGUAUAGCUCAGGGGUAGAGCAUUCGAC-UGCAGAUCGAGAGGUCCCUGGUUCAAAUCCGGGUGC---CCCCUUUGUAA--AUUUUUU-UCCCACUU--------------- -.(((((((..((((.......)))).......-....((((....))))(((((.......))))))))---)))).......--.......-........--------------- ( -30.10, z-score = -0.35, R) >droGri2.scaffold_15110 1480105 112 + 24565398 -UGGGGAUAUAGCUCAGUGGUAGAGCAUUCGAC-UGCAGAUCGAGAGGUCCCCGGUUCAAACCCGGGUGU---CCCCUAGGUAAACCUUUUUGAUGUCAAUAUUCCAAUUUAUUUCA -..(((((((.((((.......))))....(((-..((((...((.((.((((((.......))))).).---)).))(((....))).))))..))).)))))))........... ( -32.30, z-score = -0.90, R) >droAna3.scaffold_13337 22387710 90 - 23293914 -UGGGGGUAUAGCUCAGGUGUAGAGCAUUCGAC-UACAGAUCGAGAGGUCCCCGGUUCAAAUCCGGGUGC---CCCCUU-GAGU--AUCAUUU-UUUUA------------------ -.(((((((..((((.......)))).......-....((((....))))(((((.......))))))))---))))..-....--.......-.....------------------ ( -32.30, z-score = -2.27, R) >droEre2.scaffold_4784 1914658 101 + 25762168 -UGGGGGCAUAGCUCAGGGGUAGAGCGUUCGAC-UGCAGAUCGACAGGUCCCUGGUUCAAAUCCGGGUGC---CCCCUUCCUAC--ACUUUUG-CUGUGAUUUAUUUUG-------- -.(((((((..((((.......)))).......-....((((....))))(((((.......))))))))---))))......(--((.....-..)))..........-------- ( -32.10, z-score = -0.19, R) >droYak2.chr3L 1879771 104 + 24197627 -UGGGGGUAUAGCUCAGAGGUAGAGCGAUCGAU-UGCAGCUCGACAGGUCCCUGGUUCAAAUCCGGGUGC---CCCCUAGUAAACGAUUUUUUGUUUUGAUUUAUUUUG-------- -.(((((((..((((.......))))((((..(-((.....)))..))))(((((.......))))))))---))))....((((((....))))))............-------- ( -30.40, z-score = -0.89, R) >droSec1.super_2 1951139 105 + 7591821 -UGGGGGCAUAGCUCAGGGGUAGAGCGUUCGAC-UGCAGUUCGACAGGUCCCUGGUUCAAAUCCGGGUGCUGCCCCCUACUUAC--AUUUUUGGUUUUCAUUUAUUUUG-------- -.((((((...((((.......))))(((.(((-....))).))).((..(((((.......)))))..)))))))).......--.......................-------- ( -33.50, z-score = -1.11, R) >droSim1.chr3L 1480819 102 + 22553184 -UGGGGGCAUAGCUCAGGGGUAGAGCGUUCGAC-UGCAGAUCGACAGGUCCCUGGUUCAAAUCCGGGUGC---CCCCUACUUAC--AUUUUUGGCUUUCAUUUAUUUUG-------- -.(((((((..((((.......)))).......-....((((....))))(((((.......))))))))---)))).......--.......................-------- ( -30.10, z-score = -0.24, R) >consensus _UGGGGGUAUAGCUCAGGGGUAGAGCGUUCGAC_UGCAGAUCGACAGGUCCCUGGUUCAAAUCCGGGUGC___CCCCUACGUAC__AUUUUUUGUUUUAAUUUAUUUUG________ ..((((.....((((.......))))..((((........))))...(..(((((.......)))))..)...))))........................................ (-19.50 = -18.67 + -0.84)
| Location | 1,925,639 – 1,925,741 |
|---|---|
| Length | 102 |
| Sequences | 12 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 67.79 |
| Shannon entropy | 0.68227 |
| G+C content | 0.48755 |
| Mean single sequence MFE | -27.25 |
| Consensus MFE | -15.26 |
| Energy contribution | -15.76 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.29 |
| Mean z-score | -1.01 |
| Structure conservation index | 0.56 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.69 |
| SVM RNA-class probability | 0.961095 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 1925639 102 - 24543557 --------CAAAAUAAAUGAAAACCAAAAAU--GUGAGUAGGGG---GCACCCGGAUUUGAACCAGGGACCUGUCGAUCUGCA-GUCGAACGCUCUACCCCUGAGCUAUGCCCCCA- --------.......................--.......((((---((((((((.......)).))).....(((((.....-)))))..((((.......))))..))))))).- ( -30.40, z-score = -1.96, R) >anoGam1.chr2R 51280140 102 + 62725911 ---------UAAAGAGACGACGACAAAAGAGAAGUUUCUGGAG----AUGCCGGGGAUCGAACCCGGGGCCUUUCACAUGCAA-AGCGAACGCUCUACCACUGAGCUACAUCCCCU- ---------...((((((..(.........)..))))))((.(----((((((((.......)))))................-.......((((.......))))..)))).)).- ( -27.00, z-score = -0.78, R) >droMoj3.scaffold_6680 11374934 107 - 24764193 --------AAAAUUUAAUUGAAAUGAAGAAA--CCUAUAUGAGCCUGAUAGUCAAGGAAGAAUCAGCAACAUUUUUAUAGCCAUAUUUUAGACUGAGCUUUUCGAAUAACCUGUCGA --------.....(((.((((((.(.((...--...(((((.(((((((..((......))))))).............)))))))......))...).)))))).)))........ ( -10.41, z-score = 1.77, R) >droVir3.scaffold_13049 145078 109 - 25233164 -AAAAUUAAAGCAAAAGGCAACACAAAAAGU--UUACCAAGGGG---ACACCCGGGUUUGAACCGGGGACCUCUCGAUCUGCA-GUCGAAUGCUCUACCACUGAGCUAUAUCCCCA- -...............(....).........--.......((((---(..(((((.......)))))......(((((.....-)))))..((((.......))))....))))).- ( -30.10, z-score = -1.43, R) >droPer1.super_9 3419204 94 - 3637205 ---------------AAGUGGGA-AAAAAAU--GUACAAAGGGG---GCACCCGGAUUUGAACCAGGGACCUCUCGAUCUGCA-GUCGAAUGCUCUACCCCUGAGCUAUACCCCCA- ---------------........-.......--.......((((---(..(((((.......)).))).....(((((.....-)))))..((((.......))))....))))).- ( -25.10, z-score = 0.24, R) >dp4.chrXR_group6 5109246 94 - 13314419 ---------------AAGUGGGA-AAAAAAU--UUACAAAGGGG---GCACCCGGAUUUGAACCAGGGACCUCUCGAUCUGCA-GUCGAAUGCUCUACCCCUGAGCUAUACCCCCA- ---------------........-.......--.......((((---(..(((((.......)).))).....(((((.....-)))))..((((.......))))....))))).- ( -25.10, z-score = 0.09, R) >droGri2.scaffold_15110 1480105 112 - 24565398 UGAAAUAAAUUGGAAUAUUGACAUCAAAAAGGUUUACCUAGGGG---ACACCCGGGUUUGAACCGGGGACCUCUCGAUCUGCA-GUCGAAUGCUCUACCACUGAGCUAUAUCCCCA- .................(((....)))...(((((((((.(((.---...)))))))..)))))(((((....(((((.....-)))))..((((.......))))....))))).- ( -33.30, z-score = -1.63, R) >droAna3.scaffold_13337 22387710 90 + 23293914 ------------------UAAAA-AAAUGAU--ACUC-AAGGGG---GCACCCGGAUUUGAACCGGGGACCUCUCGAUCUGUA-GUCGAAUGCUCUACACCUGAGCUAUACCCCCA- ------------------.....-.......--....-..((((---(..(((((.......)))))......(((((.....-)))))..((((.......))))....))))).- ( -29.10, z-score = -2.45, R) >droEre2.scaffold_4784 1914658 101 - 25762168 --------CAAAAUAAAUCACAG-CAAAAGU--GUAGGAAGGGG---GCACCCGGAUUUGAACCAGGGACCUGUCGAUCUGCA-GUCGAACGCUCUACCCCUGAGCUAUGCCCCCA- --------.........((.(.(-((....)--)).))).((((---((((((((.......)).))).....(((((.....-)))))..((((.......))))..))))))).- ( -31.10, z-score = -1.16, R) >droYak2.chr3L 1879771 104 - 24197627 --------CAAAAUAAAUCAAAACAAAAAAUCGUUUACUAGGGG---GCACCCGGAUUUGAACCAGGGACCUGUCGAGCUGCA-AUCGAUCGCUCUACCUCUGAGCUAUACCCCCA- --------............((((........))))....((((---(..(((((.......)).)))....(((((......-.))))).((((.......))))....))))).- ( -25.80, z-score = -1.45, R) >droSec1.super_2 1951139 105 - 7591821 --------CAAAAUAAAUGAAAACCAAAAAU--GUAAGUAGGGGGCAGCACCCGGAUUUGAACCAGGGACCUGUCGAACUGCA-GUCGAACGCUCUACCCCUGAGCUAUGCCCCCA- --------.......................--.......(((((((...(((((.......)).))).....((((.(....-)))))..((((.......))))..))))))).- ( -29.20, z-score = -1.53, R) >droSim1.chr3L 1480819 102 - 22553184 --------CAAAAUAAAUGAAAGCCAAAAAU--GUAAGUAGGGG---GCACCCGGAUUUGAACCAGGGACCUGUCGAUCUGCA-GUCGAACGCUCUACCCCUGAGCUAUGCCCCCA- --------.......................--.......((((---((((((((.......)).))).....(((((.....-)))))..((((.......))))..))))))).- ( -30.40, z-score = -1.84, R) >consensus ________CAAAAUAAAUUAAAACAAAAAAU__GUAACAAGGGG___GCACCCGGAUUUGAACCAGGGACCUCUCGAUCUGCA_GUCGAACGCUCUACCCCUGAGCUAUACCCCCA_ ........................................((((......(((((.......)).))).....((((........))))..((((.......)))).....)))).. (-15.26 = -15.76 + 0.50)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:56:47 2011