
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 1,903,068 – 1,903,183 |
| Length | 115 |
| Max. P | 0.732753 |

| Location | 1,903,068 – 1,903,183 |
|---|---|
| Length | 115 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 71.11 |
| Shannon entropy | 0.54711 |
| G+C content | 0.50439 |
| Mean single sequence MFE | -33.47 |
| Consensus MFE | -16.20 |
| Energy contribution | -18.98 |
| Covariance contribution | 2.78 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.36 |
| Structure conservation index | 0.48 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.50 |
| SVM RNA-class probability | 0.721592 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 1903068 115 + 24543557 AGUGCCGGGGGAACCCUCUCCGCUGGCAGGUCGUCGUUACCUUAAGCCACAGACAAUAUGCUAAUUGGCCAACA-GCCAUUAGCU----GCCGGCACAGUUUCAUUUGCUUACUAAUUGA .((((((((....)))........((((.(((((.(((......))).)).))).....((((((.(((.....-))))))))))----))))))))....................... ( -37.60, z-score = -1.46, R) >droSim1.chr3L 1458304 111 + 22553184 AGUGCCGGGG-AACCCUCUCCGCUGGCAGGUCGU---UACCUUAAGCCACAGACAAUAUGCUAAUUGGCCAGCA-GCCACUGGCU----ACCGGCACAGUUUCAUUUGCUUACUAAUUGA .(((((((((-(......)))((((((((((...---.))))...........((((......))))))))))(-(((...))))----.)))))))....................... ( -37.80, z-score = -1.64, R) >droSec1.super_2 1928725 88 + 7591821 AGUGCCGGGG-AACCCUCUCCGCUGGCAGGUCGU---UACCUUAAGCAACAGACAAUAUGCUAAUUGGCCAGCA-GCCACUGGCU----ACCGGCAC----------------------- .(((((((((-(......)))((((((((((...---.))))..((((..........)))).....))))))(-(((...))))----.)))))))----------------------- ( -37.00, z-score = -2.84, R) >droYak2.chr3L 1856711 115 + 24197627 AGUGCCGGGG-AACCCUCUCCGCUGGCAGGUCGU---UACCUUAAGCCACAGACAAUAUGCUAAUUGGGCAGCA-GCCACUGGCUGGCUACCGGCACAGUUUCAUUUGCUUACUAAUUGA .(((((((((-(......)))((..((((((...---.)))).......(((.((((......))))(((....-))).)))))..))..)))))))....................... ( -38.10, z-score = -0.54, R) >droEre2.scaffold_4784 1890580 110 + 25762168 AGUGGCGGGG-AACCCUCUCCGCUGGCAGGUCGU---UACCUUAAGC-ACAGACAAUAUGCUAAUUGGGCAGCA-ACCACCGGCU----ACCGGCACAGUUUCAUUUGCUUACUAAUUGA .(((.(((((-(......)))(((((..(((.((---(.(((..(((-(.(.....).))))....))).))).-))).))))).----.))).)))....................... ( -32.80, z-score = -0.39, R) >droAna3.scaffold_13337 22359880 91 - 23293914 ---------------------------AAUGCAGGGGAACCCUGCAUUGCAAA-AAUAUGCAAAUAUGCUAAUAAGCUACCUGCUACC-ACCGACACAGUUUCAAUUUAUAACUAAUUAA ---------------------------..(((((((...)))))))(((((..-....)))))...........(((.....)))...-........((((.........))))...... ( -17.50, z-score = -1.29, R) >consensus AGUGCCGGGG_AACCCUCUCCGCUGGCAGGUCGU___UACCUUAAGCCACAGACAAUAUGCUAAUUGGCCAGCA_GCCACUGGCU____ACCGGCACAGUUUCAUUUGCUUACUAAUUGA .(((((((.............(((...((((.......))))..)))............(((...((((......))))..)))......)))))))....................... (-16.20 = -18.98 + 2.78)

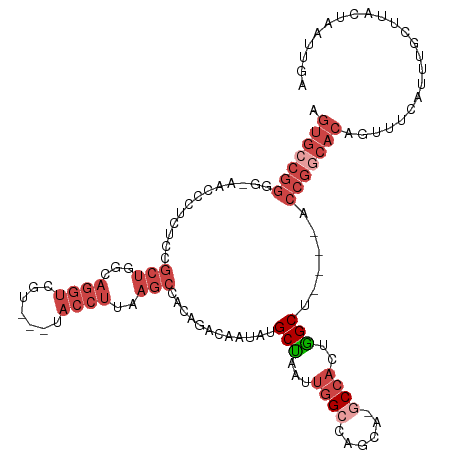
| Location | 1,903,068 – 1,903,183 |
|---|---|
| Length | 115 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 71.11 |
| Shannon entropy | 0.54711 |
| G+C content | 0.50439 |
| Mean single sequence MFE | -36.00 |
| Consensus MFE | -18.42 |
| Energy contribution | -20.65 |
| Covariance contribution | 2.23 |
| Combinations/Pair | 1.29 |
| Mean z-score | -1.24 |
| Structure conservation index | 0.51 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.53 |
| SVM RNA-class probability | 0.732753 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 1903068 115 - 24543557 UCAAUUAGUAAGCAAAUGAAACUGUGCCGGC----AGCUAAUGGC-UGUUGGCCAAUUAGCAUAUUGUCUGUGGCUUAAGGUAACGACGACCUGCCAGCGGAGAGGGUUCCCCCGGCACU .......................(((((((.----.(((((((((-.....))).))))))....(.((((((((...((((.......)))))))).)))).)((....))))))))). ( -39.70, z-score = -1.30, R) >droSim1.chr3L 1458304 111 - 22553184 UCAAUUAGUAAGCAAAUGAAACUGUGCCGGU----AGCCAGUGGC-UGCUGGCCAAUUAGCAUAUUGUCUGUGGCUUAAGGUA---ACGACCUGCCAGCGGAGAGGGUU-CCCCGGCACU .......................(((((((.----((((.....(-(((((((.....(((.(((.....))))))..((((.---...))))))))))))....))))-..))))))). ( -38.90, z-score = -0.91, R) >droSec1.super_2 1928725 88 - 7591821 -----------------------GUGCCGGU----AGCCAGUGGC-UGCUGGCCAAUUAGCAUAUUGUCUGUUGCUUAAGGUA---ACGACCUGCCAGCGGAGAGGGUU-CCCCGGCACU -----------------------(((((((.----((((.....(-(((((((((((..((.....))..))))....((((.---...))))))))))))....))))-..))))))). ( -40.00, z-score = -2.65, R) >droYak2.chr3L 1856711 115 - 24197627 UCAAUUAGUAAGCAAAUGAAACUGUGCCGGUAGCCAGCCAGUGGC-UGCUGCCCAAUUAGCAUAUUGUCUGUGGCUUAAGGUA---ACGACCUGCCAGCGGAGAGGGUU-CCCCGGCACU .......................(((((((((((.((((...)))-)))))))............(.((((((((...((((.---...)))))))).)))).)(((..-.)))))))). ( -39.70, z-score = -0.83, R) >droEre2.scaffold_4784 1890580 110 - 25762168 UCAAUUAGUAAGCAAAUGAAACUGUGCCGGU----AGCCGGUGGU-UGCUGCCCAAUUAGCAUAUUGUCUGU-GCUUAAGGUA---ACGACCUGCCAGCGGAGAGGGUU-CCCCGCCACU .......................(((.(((.----.((.((((((-((.((((.....((((((.....)))-)))...))))---.))))).))).))((((....))-))))).))). ( -32.80, z-score = -0.09, R) >droAna3.scaffold_13337 22359880 91 + 23293914 UUAAUUAGUUAUAAAUUGAAACUGUGUCGGU-GGUAGCAGGUAGCUUAUUAGCAUAUUUGCAUAUU-UUUGCAAUGCAGGGUUCCCCUGCAUU--------------------------- .....(((((.........)))))((.(((.-.(((((((((((((....))).))))))).))).-.)))))((((((((...)))))))).--------------------------- ( -24.90, z-score = -1.63, R) >consensus UCAAUUAGUAAGCAAAUGAAACUGUGCCGGU____AGCCAGUGGC_UGCUGGCCAAUUAGCAUAUUGUCUGUGGCUUAAGGUA___ACGACCUGCCAGCGGAGAGGGUU_CCCCGGCACU .......................(((((((......((((((.....))))))...........((.(((((......((((.......))))....))))).)).......))))))). (-18.42 = -20.65 + 2.23)

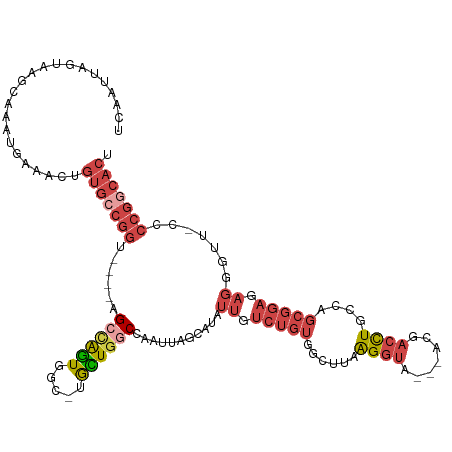
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:56:41 2011