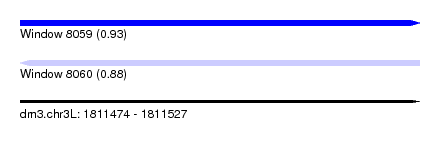
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 1,811,474 – 1,811,527 |
| Length | 53 |
| Max. P | 0.926888 |

| Location | 1,811,474 – 1,811,527 |
|---|---|
| Length | 53 |
| Sequences | 5 |
| Columns | 53 |
| Reading direction | forward |
| Mean pairwise identity | 94.71 |
| Shannon entropy | 0.09112 |
| G+C content | 0.33070 |
| Mean single sequence MFE | -7.68 |
| Consensus MFE | -7.42 |
| Energy contribution | -7.62 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.66 |
| Structure conservation index | 0.97 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.33 |
| SVM RNA-class probability | 0.926888 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
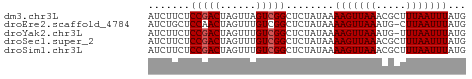
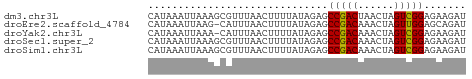
>dm3.chr3L 1811474 53 + 24543557 AUCUUCUCCGACUAGUUAGUCGGCUCUAUAAAAGUUAAACGCUUUAAUUUAUG .......((((((....)))))).......(((((.....)))))........ ( -9.20, z-score = -2.06, R) >droEre2.scaffold_4784 1795976 52 + 25762168 AUCUGCUCCAACUAGUUUGUCGGCUCUAUAAAAGUUAAAUG-CUUAAUUUAUG ....(((.(((.....)))..))).......(((((((...-.)))))))... ( -3.00, z-score = 0.82, R) >droYak2.chr3L 1763721 52 + 24197627 AUCUUCUCCGACUAGUUUGUCGGCUCUAUAAAAGUUAAAUG-UUUAAUUUAUG .......(((((......)))))........(((((((...-.)))))))... ( -8.60, z-score = -2.58, R) >droSec1.super_2 1837396 53 + 7591821 AUCUUCUCCGACUAGUUUGUCGGCUCUAUAAAAGUUAAACGCUUUAAUUUAUG .......(((((......))))).......(((((.....)))))........ ( -8.80, z-score = -2.25, R) >droSim1.chr3L 1367594 53 + 22553184 AUCUUCUCCGACUAGUUUGUCGGCUCUAUAAAAGUUAAACGCUUUAAUUUAUG .......(((((......))))).......(((((.....)))))........ ( -8.80, z-score = -2.25, R) >consensus AUCUUCUCCGACUAGUUUGUCGGCUCUAUAAAAGUUAAACGCUUUAAUUUAUG .......(((((......)))))........(((((((.....)))))))... ( -7.42 = -7.62 + 0.20)
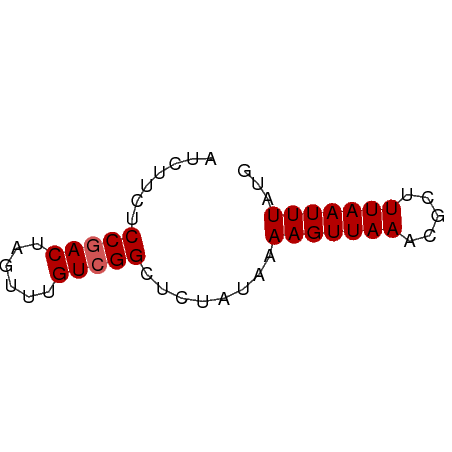
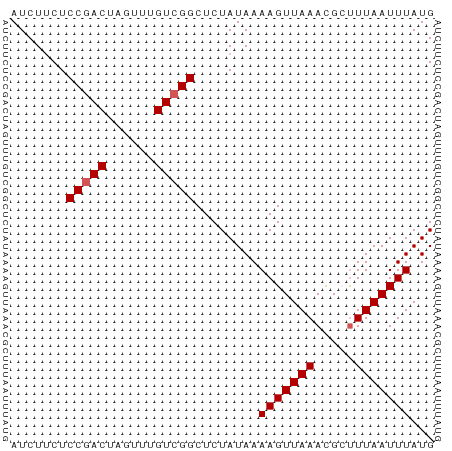
| Location | 1,811,474 – 1,811,527 |
|---|---|
| Length | 53 |
| Sequences | 5 |
| Columns | 53 |
| Reading direction | reverse |
| Mean pairwise identity | 94.71 |
| Shannon entropy | 0.09112 |
| G+C content | 0.33070 |
| Mean single sequence MFE | -8.08 |
| Consensus MFE | -7.48 |
| Energy contribution | -7.32 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.20 |
| Mean z-score | -1.75 |
| Structure conservation index | 0.93 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.05 |
| SVM RNA-class probability | 0.882220 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
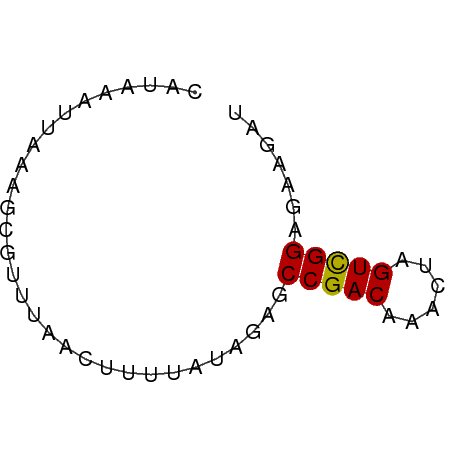
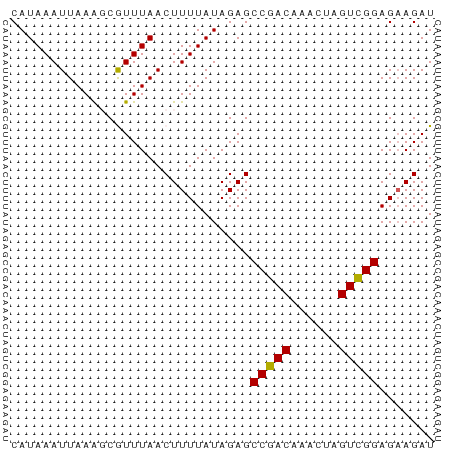
>dm3.chr3L 1811474 53 - 24543557 CAUAAAUUAAAGCGUUUAACUUUUAUAGAGCCGACUAACUAGUCGGAGAAGAU ...................(((((......((((((....))))))))))).. ( -8.90, z-score = -1.47, R) >droEre2.scaffold_4784 1795976 52 - 25762168 CAUAAAUUAAG-CAUUUAACUUUUAUAGAGCCGACAAACUAGUUGGAGCAGAU ...........-...............(..(((((......)))))..).... ( -7.50, z-score = -1.59, R) >droYak2.chr3L 1763721 52 - 24197627 CAUAAAUUAAA-CAUUUAACUUUUAUAGAGCCGACAAACUAGUCGGAGAAGAU ...........-.......(((((......(((((......)))))))))).. ( -8.00, z-score = -2.69, R) >droSec1.super_2 1837396 53 - 7591821 CAUAAAUUAAAGCGUUUAACUUUUAUAGAGCCGACAAACUAGUCGGAGAAGAU ...................(((((......(((((......)))))))))).. ( -8.00, z-score = -1.51, R) >droSim1.chr3L 1367594 53 - 22553184 CAUAAAUUAAAGCGUUUAACUUUUAUAGAGCCGACAAACUAGUCGGAGAAGAU ...................(((((......(((((......)))))))))).. ( -8.00, z-score = -1.51, R) >consensus CAUAAAUUAAAGCGUUUAACUUUUAUAGAGCCGACAAACUAGUCGGAGAAGAU ..............................(((((......)))))....... ( -7.48 = -7.32 + -0.16)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:56:35 2011