| Sequence ID | dm3.chr3L |
|---|---|
| Location | 1,784,560 – 1,784,655 |
| Length | 95 |
| Max. P | 0.803410 |

| Location | 1,784,560 – 1,784,655 |
|---|---|
| Length | 95 |
| Sequences | 8 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 70.44 |
| Shannon entropy | 0.57148 |
| G+C content | 0.46451 |
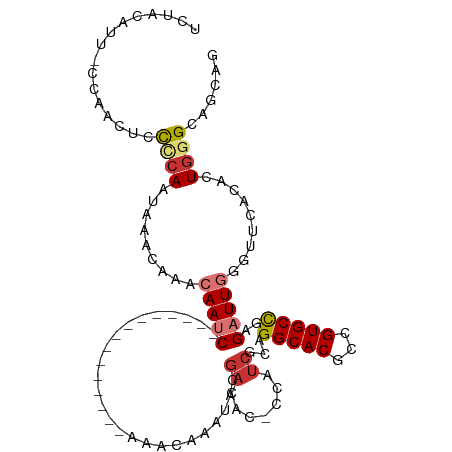
| Mean single sequence MFE | -20.87 |
| Consensus MFE | -8.02 |
| Energy contribution | -7.71 |
| Covariance contribution | -0.31 |
| Combinations/Pair | 1.36 |
| Mean z-score | -1.87 |
| Structure conservation index | 0.38 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.74 |
| SVM RNA-class probability | 0.803410 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 1784560 95 - 24543557 UCUACAUU-CCAACUCCCCAAUAAACAAUCAAUC---------------AAACAAAUACGACAC-CCAUCGCAGGCACGCCGUGCCGAGAUUGGGUUCACAUUGGGCAUCAG ........-.......((((((......((((((---------------.........(((...-...)))..(((((...)))))..))))))......))))))...... ( -21.30, z-score = -2.39, R) >droSim1.chr3L 1340940 95 - 22553184 UCUACAUU-CCAACUCCUCAAUAAACAAUCAAUC---------------AAACAAAUACGACAC-CUAUCGCAGGCACGCCGUGCCGAGAUUGGGUUCACAUUGGGCAUCAG ........-.......((((((......((((((---------------.........(((...-...)))..(((((...)))))..))))))......))))))...... ( -18.30, z-score = -1.63, R) >droSec1.super_2 1810750 95 - 7591821 UCUACAUU-CCAACUCCCCAAUAAACAAUCAAUC---------------AAACAAAUACGACAC-CCAUCGCAGGCACGCCGUGCCGAGAUUGGGUUCACAUUGGGCAUCAG ........-.......((((((......((((((---------------.........(((...-...)))..(((((...)))))..))))))......))))))...... ( -21.30, z-score = -2.39, R) >droYak2.chr3L 1736886 95 - 24197627 UCUACAUU-CCAACUCCCCAAUAAACAAACAAUC---------------AAACAAAUACGAUAC-CCAUCACAGGCACGCCGUGCCGAGAUUGGGUUCACCCUGGGCAGCAG ........-....((.((((..........((((---------------..........(((..-..)))...(((((...)))))..))))(((....))))))).))... ( -20.10, z-score = -1.89, R) >droEre2.scaffold_4784 1769251 95 - 25762168 UCUACAUU-CCAACUCCCCAAUAAACAAACAAUC---------------AAACAAAUACGAUAC-CCAUCGCAGGCACGCCGUGCCGAGAUUGGUUUCACACUGGGCAGCAG ........-....((.((((.........(((((---------------.........((((..-..))))..(((((...)))))..))))).........)))).))... ( -20.37, z-score = -2.28, R) >droAna3.scaffold_13337 22240854 111 + 23293914 UCUGCAUU-CCGACACUCAAAUGAAAAUACAACCAACAACAUAUAUAUGAAACACGUGCGACACAAUCUCGCAGGCACACCGUGCUGAGAUUGAAUUUACACUGGGCAGCAG .((((..(-(((...........................(((....)))......(((..(..((((((((((((....)).))).)))))))...)..)))))))..)))) ( -23.00, z-score = -1.48, R) >dp4.chrXR_group8 526816 105 - 9212921 UCUGCAUUACAAACAUCACAACCCAAACAAAAUCCGAAUCG---GUAUCCUGUAUCCUGGAUCCUGGAUC-UAGGCACAACGUGCUGAGAUUGAGUUCACUCUGGGCAG--- .....................((((.....((((....(((---((((..(((..((((((((...))))-)))).)))..)))))))))))(((....)))))))...--- ( -26.40, z-score = -1.34, R) >droWil1.scaffold_180727 1502461 78 + 2741493 --------------UCUGCAUUUAAAAUAAAAAC--------------CAAAUACAUUUCACAC------UCAGGCACAACGUGCUGAGAUUGAGUUUACUCUGGGCAGCAG --------------.((((.((((...))))..(--------------((.............(------((((.(((...))))))))...(((....))))))))))... ( -16.20, z-score = -1.57, R) >consensus UCUACAUU_CCAACUCCCCAAUAAACAAACAAUC_______________AAACAAAUACGACAC_CCAUCGCAGGCACGCCGUGCCGAGAUUGGGUUCACACUGGGCAGCAG ................((((.....................................................(((((...)))))..((......))....))))...... ( -8.02 = -7.71 + -0.31)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:56:34 2011