
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 1,781,356 – 1,781,519 |
| Length | 163 |
| Max. P | 0.761845 |

| Location | 1,781,356 – 1,781,475 |
|---|---|
| Length | 119 |
| Sequences | 15 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 88.50 |
| Shannon entropy | 0.27285 |
| G+C content | 0.52241 |
| Mean single sequence MFE | -40.05 |
| Consensus MFE | -26.57 |
| Energy contribution | -26.55 |
| Covariance contribution | -0.02 |
| Combinations/Pair | 1.33 |
| Mean z-score | -2.08 |
| Structure conservation index | 0.66 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.61 |
| SVM RNA-class probability | 0.761845 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 1781356 119 - 24543557 GAAAGCUUCGGCCUUCAAUUCUUGGUUCGAAAAUGCCGAGGAGGAUCUCACAGAUCCUGUUCGCUGCAAUUCGAUCGAAGAAAUCCGCGCCCUGCGCGACGCCCAUGCCCAAUUCCAGG- (((.((....)).)))..(((((.(.(((((..(((((((.((((((.....)))))).))))..))).))))).).)))))...(((((...))))).....................- ( -40.80, z-score = -2.09, R) >anoGam1.chr2L 12665581 119 + 48795086 GAAGGCGUCGGCAUUCAACUCGUGGUUCGAGAACGCGGAGGAAGAUCUGACCGAUCCGGUGCGCUGCAACUCGAUCGAGGAGAUCAAGGCGCUGCGGGAGGCACAUGCCGCGUUCCAGG- (((.(((((((..(((...(((((.........)))))..)))...)))))...(((.(((((((.(..(((....)))..).....))))).)).)))(((....))))).)))....- ( -40.60, z-score = 1.17, R) >droGri2.scaffold_15110 413702 119 - 24565398 GAAAGCUUCGGCCUUCAAUUCUUGGUUCGAGAAUGCCGAAGAGGAUCUCACAGAUCCUGUUCGCUGCAAUUCGAUCGAAGAAAUACGCGCCUUGCGUGAUGCCCACGCUCAAUUCCAAG- (((.((....)).)))..(((((.(.(((((..(((((((.((((((.....)))))).))))..))).))))).).)))))...........(((((.....)))))...........- ( -40.70, z-score = -2.78, R) >droVir3.scaffold_10322 371446 119 + 456481 GAAAGCUUCGGCCUUCAAUUCUUGGUUCGAAAAUGCCGAGGAGGAUCUCACGGAUCCUGUUCGCUGCAAUUCGAUCGAAGAAAUACGCGCCCUGCGUGACGCGCACGCACAAUUCCAAG- (((.((....)).)))..(((((.(.(((((..(((((((.((((((.....)))))).))))..))).))))).).)))))....(((...((((.....)))))))...........- ( -42.20, z-score = -2.44, R) >droMoj3.scaffold_6680 22621680 119 + 24764193 GAAAGCUUCGGCCUUCAAUUCUUGGUUCGAAAAUGCCGAGGAGGAUCUCACAGAUCCUGUUCGCUGCAAUUCGAUCGAAGAAAUACGCGCGCUGCGUGAUGCCCACGCCCAAUUCCAAG- (((.((....)).)))..(((((.(.(((((..(((((((.((((((.....)))))).))))..))).))))).).)))))...........(((((.....)))))...........- ( -40.30, z-score = -2.34, R) >droWil1.scaffold_180727 1498654 119 + 2741493 GAAAGCUUCGGCCUUCAAUUCUUGGUUCGAAAAUGCCGAAGAGGAUCUCACAGAUCCUGUUCGCUGCAAUUCGAUCGAAGAAAUACGCGCCCUGCGCGACGCCCAUGCACAAUUCCAAG- ....((...(((......(((((.(.(((((..(((((((.((((((.....)))))).))))..))).))))).).)))))...(((((...)))))..)))...))...........- ( -42.00, z-score = -3.67, R) >droPer1.super_32 33410 119 + 992631 GAAAGCUUCGGCCUUCAAUUCUUGGUUCGAAAAUGCCGAAGAGGAUCUCACAGAUCCUGUUCGCUGCAAUUCGAUCGAAGAAAUACGCGCGUUGCGCGACGCCCACGCACAAUUCCAAG- (((.((....)).)))..(((((.(.(((((..(((((((.((((((.....)))))).))))..))).))))).).)))))...(((((...)))))..((....))...........- ( -42.00, z-score = -3.13, R) >dp4.chrXR_group8 522706 119 - 9212921 GAAAGCUUCGGCCUUCAAUUCUUGGUUCGAAAAUGCCGAAGAGGAUCUCACAGAUCCUGUUCGCUGCAAUUCGAUCGAAGAAAUACGCGCGUUGCGCGACGCCCACGCACAAUUCCAAG- (((.((....)).)))..(((((.(.(((((..(((((((.((((((.....)))))).))))..))).))))).).)))))...(((((...)))))..((....))...........- ( -42.00, z-score = -3.13, R) >droAna3.scaffold_13337 22237798 119 + 23293914 GAAAGCUUCGGCCUUCAAUUCUUGGUUCGAGAAUGCCGAAGAGGAUCUCACAGAUCCUGUUCGCUGCAACUCGAUCGAAGAAAUUCGCGCCCUGCGCGACGCCCACGCCCAAUUCCAGG- (((.((....)).)))..(((((.(.(((((..(((((((.((((((.....)))))).))))..))).))))).).)))))..((((((...))))))....................- ( -44.20, z-score = -3.13, R) >droEre2.scaffold_4784 1766057 119 - 25762168 GAAAGCUUCGGCCUUCAAUUCUUGGUUCGAAAAUGCCGAGGAGGAUCUUACAGAUCCUGUUCGCUGCAAUUCGAUCGAAGAAAUCCGCGCCCUGCGCGACGCCCAUGCCCAAUUCCAGG- (((.((....)).)))..(((((.(.(((((..(((((((.((((((.....)))))).))))..))).))))).).)))))...(((((...))))).....................- ( -40.80, z-score = -2.24, R) >droYak2.chr3L 1733755 119 - 24197627 GAAAGCUUCGGCCUUCAAUUCUUGGUUCGAAAAUGCCGAGGAGGAUCUCACAGAUCCUGUUCGCUGCAAUUCGAUCGAAGAAAUUCGCGCCCUGCGCGACGCCCAUGCCCAAUUCCAGG- (((.((....)).)))..(((((.(.(((((..(((((((.((((((.....)))))).))))..))).))))).).)))))..((((((...))))))....................- ( -42.00, z-score = -2.39, R) >droSec1.super_2 1807555 119 - 7591821 GAAAGCUUCGGCCUUCAAUUCUUGGUUCGAAAAUGCCGAGGAGGAUCUCACAGAUCCUGUUCGCUGCAAUUCGAUCGAAGAAAUCCGCGCCCUGCGCGACGCCCAUGCCCAAUUCCAGG- (((.((....)).)))..(((((.(.(((((..(((((((.((((((.....)))))).))))..))).))))).).)))))...(((((...))))).....................- ( -40.80, z-score = -2.09, R) >droSim1.chr3L 1337775 119 - 22553184 GAAAGCUUCGGCCUUCAAUUCUUGGUUCGAAAAUGCCGAGGAGGAUCUCACAGAUCCUGUUCGCUGCAAUUCGAUCGAAGAAAUCCGCGCCCUGCGCGACGCCCAUGCCCAAUUCCAGG- (((.((....)).)))..(((((.(.(((((..(((((((.((((((.....)))))).))))..))).))))).).)))))...(((((...))))).....................- ( -40.80, z-score = -2.09, R) >apiMel3.Group3 1562978 119 + 12341916 GAAAGCAUCUGCUUUCAAUUCAUGGUUUGAAAAUGCAGAAGAAGAUUUAACAGAUCCUGUACGAUGUAAUAGUAUUGAAGAAAUUCGAGCUCUCAGAGAAGCACAUGCACAAUUUCAAG- ....((((.((((((........((((((((..(((((.....((((.....)))))))))(((((......)))))......))))))))......)))))).))))...........- ( -23.74, z-score = -0.02, R) >triCas2.ChLG2 749377 120 - 12900155 GAGAGCGUCUGCGUUCAAUUCUUGGUUUGAAAAUGCUGAGGAGGAUUUGACCGAUCCAGUGCGAUGCAAUUCCAUUGAGGAGAUCCGAGCGUUGAGAGAGGCUCAUGCACAAUUCCAGGU ..((((.(((((((((..((((..((.(....).))..))))((((((..((....(((((.((......))))))).))))))))))))))....))).))))................ ( -37.80, z-score = -0.83, R) >consensus GAAAGCUUCGGCCUUCAAUUCUUGGUUCGAAAAUGCCGAGGAGGAUCUCACAGAUCCUGUUCGCUGCAAUUCGAUCGAAGAAAUACGCGCCCUGCGCGACGCCCAUGCCCAAUUCCAGG_ (((.((....)).)))..(((((.(.(((((..(((((.(.((((((.....)))))).).))..))).))))).).)))))....(.((...((.....))....)).).......... (-26.57 = -26.55 + -0.02)
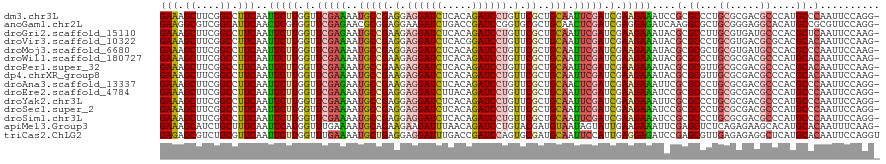
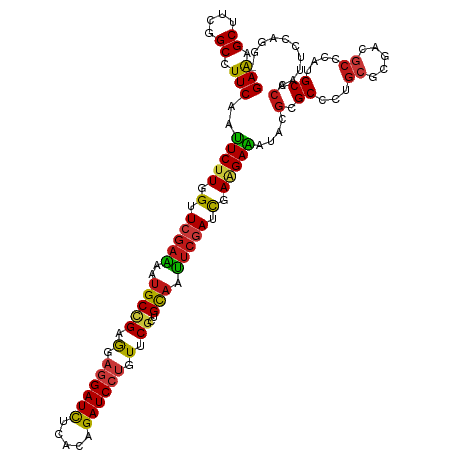
| Location | 1,781,395 – 1,781,515 |
|---|---|
| Length | 120 |
| Sequences | 15 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 91.06 |
| Shannon entropy | 0.21504 |
| G+C content | 0.46833 |
| Mean single sequence MFE | -37.81 |
| Consensus MFE | -26.93 |
| Energy contribution | -26.89 |
| Covariance contribution | -0.05 |
| Combinations/Pair | 1.29 |
| Mean z-score | -2.00 |
| Structure conservation index | 0.71 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.33 |
| SVM RNA-class probability | 0.647722 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 1781395 120 - 24543557 AGUUCCGCCAGAUCGAGGAACUCUACUUGACAUUUGCCAAGAAAGCUUCGGCCUUCAAUUCUUGGUUCGAAAAUGCCGAGGAGGAUCUCACAGAUCCUGUUCGCUGCAAUUCGAUCGAAG ((((((..........))))))....((((.....((((((((.((....))......))))))))(((((..(((((((.((((((.....)))))).))))..))).))))))))).. ( -39.00, z-score = -1.98, R) >anoGam1.chr2L 12665620 120 + 48795086 AGUUCCGCCAGAUCGAGGAUCUGUAUCUGACGUUCGCCAAGAAGGCGUCGGCAUUCAACUCGUGGUUCGAGAACGCGGAGGAAGAUCUGACCGAUCCGGUGCGCUGCAACUCGAUCGAGG (((..((((.(((((.((((((...((((.((((((((.....))).((((...(((.....))).)))))))))))))...))))))...))))).)))).)))....(((....))). ( -43.50, z-score = -0.26, R) >droGri2.scaffold_15110 413741 120 - 24565398 AAUUCCGCCAAAUCGAGGAACUCUACUUGACAUUUGCCAAGAAAGCUUCGGCCUUCAAUUCUUGGUUCGAGAAUGCCGAAGAGGAUCUCACAGAUCCUGUUCGCUGCAAUUCGAUCGAAG ............((((.(((.....(((((.....((((((((.((....))......)))))))))))))..(((((((.((((((.....)))))).))))..))).)))..)))).. ( -37.20, z-score = -2.19, R) >droVir3.scaffold_10322 371485 120 + 456481 AGUUCCGCCAAAUCGAGGAACUCUACUUGACAUUUGCCAAGAAAGCUUCGGCCUUCAAUUCUUGGUUCGAAAAUGCCGAGGAGGAUCUCACGGAUCCUGUUCGCUGCAAUUCGAUCGAAG ((((((..........))))))....((((.....((((((((.((....))......))))))))(((((..(((((((.((((((.....)))))).))))..))).))))))))).. ( -39.10, z-score = -2.11, R) >droMoj3.scaffold_6680 22621719 120 + 24764193 AAUUCCGCCAAAUCGAGGAACUCUAUUUGACAUUUGCCAAGAAAGCUUCGGCCUUCAAUUCUUGGUUCGAAAAUGCCGAGGAGGAUCUCACAGAUCCUGUUCGCUGCAAUUCGAUCGAAG ..((((..........)))).....(((((.....((((((((.((....))......))))))))(((((..(((((((.((((((.....)))))).))))..))).)))))))))). ( -36.50, z-score = -1.96, R) >droWil1.scaffold_180727 1498693 120 + 2741493 AAUUCCGCCAAAUCGAGGAACUCUACUUGACAUUUGCCAAGAAAGCUUCGGCCUUCAAUUCUUGGUUCGAAAAUGCCGAAGAGGAUCUCACAGAUCCUGUUCGCUGCAAUUCGAUCGAAG ..(((.(((((.(((((........)))))..........(((.((....)).))).....)))))(((((..(((((((.((((((.....)))))).))))..))).)))))..))). ( -36.60, z-score = -2.43, R) >droPer1.super_32 33449 120 + 992631 AGUUCCGCCAAAUCGAGGAACUCUACUUGACAUUUGCCAAGAAAGCUUCGGCCUUCAAUUCUUGGUUCGAAAAUGCCGAAGAGGAUCUCACAGAUCCUGUUCGCUGCAAUUCGAUCGAAG ((((((..........))))))....((((.....((((((((.((....))......))))))))(((((..(((((((.((((((.....)))))).))))..))).))))))))).. ( -39.30, z-score = -2.90, R) >dp4.chrXR_group8 522745 120 - 9212921 AGUUCCGCCAAAUCGAGGAACUCUACUUGACAUUUGCCAAGAAAGCUUCGGCCUUCAAUUCUUGGUUCGAAAAUGCCGAAGAGGAUCUCACAGAUCCUGUUCGCUGCAAUUCGAUCGAAG ((((((..........))))))....((((.....((((((((.((....))......))))))))(((((..(((((((.((((((.....)))))).))))..))).))))))))).. ( -39.30, z-score = -2.90, R) >droAna3.scaffold_13337 22237837 120 + 23293914 AGUUCCGCCAGAUCGAGGAACUCUACUUGACAUUUGCCAAGAAAGCUUCGGCCUUCAAUUCUUGGUUCGAGAAUGCCGAAGAGGAUCUCACAGAUCCUGUUCGCUGCAACUCGAUCGAAG ..........(((((((........(((((.....((((((((.((....))......)))))))))))))..(((((((.((((((.....)))))).))))..))).))))))).... ( -43.90, z-score = -3.30, R) >droEre2.scaffold_4784 1766096 120 - 25762168 AGUUCCGCCAGAUCGAGGAACUCUACUUGACAUUUGCCAAGAAAGCUUCGGCCUUCAAUUCUUGGUUCGAAAAUGCCGAGGAGGAUCUUACAGAUCCUGUUCGCUGCAAUUCGAUCGAAG ((((((..........))))))....((((.....((((((((.((....))......))))))))(((((..(((((((.((((((.....)))))).))))..))).))))))))).. ( -39.00, z-score = -2.19, R) >droYak2.chr3L 1733794 120 - 24197627 AGUUCCGCCAGAUCGAGGAACUCUACUUGACAUUUGCCAAGAAAGCUUCGGCCUUCAAUUCUUGGUUCGAAAAUGCCGAGGAGGAUCUCACAGAUCCUGUUCGCUGCAAUUCGAUCGAAG ((((((..........))))))....((((.....((((((((.((....))......))))))))(((((..(((((((.((((((.....)))))).))))..))).))))))))).. ( -39.00, z-score = -1.98, R) >droSec1.super_2 1807594 120 - 7591821 AGUUCCGCCAGAUCGAGGAACUCUACUUGACAUUUGCCAAGAAAGCUUCGGCCUUCAAUUCUUGGUUCGAAAAUGCCGAGGAGGAUCUCACAGAUCCUGUUCGCUGCAAUUCGAUCGAAG ((((((..........))))))....((((.....((((((((.((....))......))))))))(((((..(((((((.((((((.....)))))).))))..))).))))))))).. ( -39.00, z-score = -1.98, R) >droSim1.chr3L 1337814 120 - 22553184 AGUUCCGCCAGAUCGAGGAACUCUACUUGACAUUUGCCAAGAAAGCUUCGGCCUUCAAUUCUUGGUUCGAAAAUGCCGAGGAGGAUCUCACAGAUCCUGUUCGCUGCAAUUCGAUCGAAG ((((((..........))))))....((((.....((((((((.((....))......))))))))(((((..(((((((.((((((.....)))))).))))..))).))))))))).. ( -39.00, z-score = -1.98, R) >apiMel3.Group3 1563017 120 + 12341916 AAUUCAGACAAAUUGAAGAAUUGUAUUUAACAUUUGCCAAGAAAGCAUCUGCUUUCAAUUCAUGGUUUGAAAAUGCAGAAGAAGAUUUAACAGAUCCUGUACGAUGUAAUAGUAUUGAAG ..(((((((....((((....(((.....)))........((((((....))))))..))))..)))))))..(((((.....((((.....)))))))))(((((......)))))... ( -22.10, z-score = -0.55, R) >triCas2.ChLG2 749417 120 - 12900155 AAUUCCGACAAAUCGAAGAUCUUUUCCUCACUUUUGCCAAGAGAGCGUCUGCGUUCAAUUCUUGGUUUGAAAAUGCUGAGGAGGAUUUGACCGAUCCAGUGCGAUGCAAUUCCAUUGAGG ....((.....((((.((((((..(((((((((((((((((((((((....)))))...)))))))...)))).).))))))))))))...)))).(((((.((......))))))).)) ( -34.70, z-score = -1.31, R) >consensus AGUUCCGCCAAAUCGAGGAACUCUACUUGACAUUUGCCAAGAAAGCUUCGGCCUUCAAUUCUUGGUUCGAAAAUGCCGAGGAGGAUCUCACAGAUCCUGUUCGCUGCAAUUCGAUCGAAG ............((((...................((((((((.((....))......))))))))(((((..(((((.(.((((((.....)))))).).))..))).))))))))).. (-26.93 = -26.89 + -0.05)
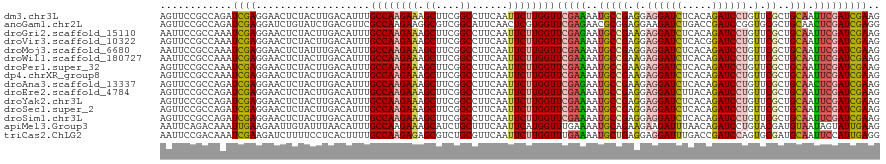
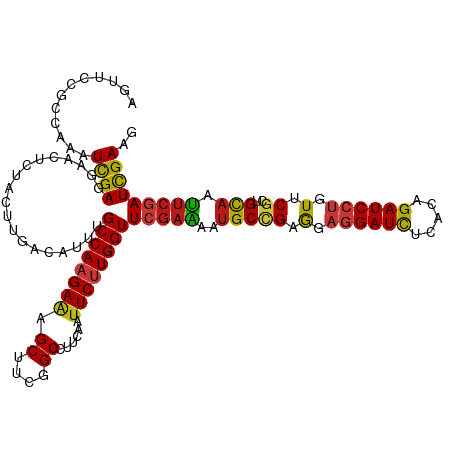
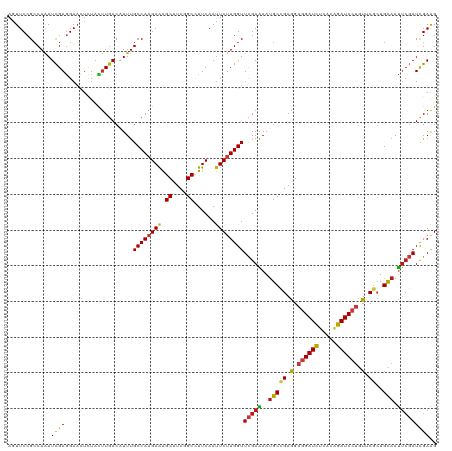
| Location | 1,781,399 – 1,781,519 |
|---|---|
| Length | 120 |
| Sequences | 15 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 90.97 |
| Shannon entropy | 0.21786 |
| G+C content | 0.47500 |
| Mean single sequence MFE | -37.77 |
| Consensus MFE | -26.32 |
| Energy contribution | -26.39 |
| Covariance contribution | 0.08 |
| Combinations/Pair | 1.31 |
| Mean z-score | -1.92 |
| Structure conservation index | 0.70 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.12 |
| SVM RNA-class probability | 0.551973 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 1781399 120 - 24543557 GAGCAGUUCCGCCAGAUCGAGGAACUCUACUUGACAUUUGCCAAGAAAGCUUCGGCCUUCAAUUCUUGGUUCGAAAAUGCCGAGGAGGAUCUCACAGAUCCUGUUCGCUGCAAUUCGAUC .((.((((((..........))))))))...........((((((((.((....))......))))))))(((((..(((((((.((((((.....)))))).))))..))).))))).. ( -39.50, z-score = -1.97, R) >anoGam1.chr2L 12665624 120 + 48795086 GACCAGUUCCGCCAGAUCGAGGAUCUGUAUCUGACGUUCGCCAAGAAGGCGUCGGCAUUCAACUCGUGGUUCGAGAACGCGGAGGAAGAUCUGACCGAUCCGGUGCGCUGCAACUCGAUC ...((((..((((.(((((.((((((...((((.((((((((.....))).((((...(((.....))).)))))))))))))...))))))...))))).)))).)))).......... ( -43.40, z-score = -0.59, R) >droGri2.scaffold_15110 413745 120 - 24565398 GAGCAAUUCCGCCAAAUCGAGGAACUCUACUUGACAUUUGCCAAGAAAGCUUCGGCCUUCAAUUCUUGGUUCGAGAAUGCCGAAGAGGAUCUCACAGAUCCUGUUCGCUGCAAUUCGAUC (.((......)))..((((((........(((((.....((((((((.((....))......)))))))))))))..(((((((.((((((.....)))))).))))..))).)))))). ( -37.50, z-score = -2.10, R) >droVir3.scaffold_10322 371489 120 + 456481 GAGCAGUUCCGCCAAAUCGAGGAACUCUACUUGACAUUUGCCAAGAAAGCUUCGGCCUUCAAUUCUUGGUUCGAAAAUGCCGAGGAGGAUCUCACGGAUCCUGUUCGCUGCAAUUCGAUC .((.((((((..........))))))))...........((((((((.((....))......))))))))(((((..(((((((.((((((.....)))))).))))..))).))))).. ( -39.60, z-score = -2.11, R) >droMoj3.scaffold_6680 22621723 120 + 24764193 GAGCAAUUCCGCCAAAUCGAGGAACUCUAUUUGACAUUUGCCAAGAAAGCUUCGGCCUUCAAUUCUUGGUUCGAAAAUGCCGAGGAGGAUCUCACAGAUCCUGUUCGCUGCAAUUCGAUC ..((......))(((((.(((...))).)))))......((((((((.((....))......))))))))(((((..(((((((.((((((.....)))))).))))..))).))))).. ( -36.20, z-score = -1.75, R) >droWil1.scaffold_180727 1498697 120 + 2741493 GAGCAAUUCCGCCAAAUCGAGGAACUCUACUUGACAUUUGCCAAGAAAGCUUCGGCCUUCAAUUCUUGGUUCGAAAAUGCCGAAGAGGAUCUCACAGAUCCUGUUCGCUGCAAUUCGAUC (.((......)))...(((((........))))).....((((((((.((....))......))))))))(((((..(((((((.((((((.....)))))).))))..))).))))).. ( -36.90, z-score = -2.37, R) >droPer1.super_32 33453 120 + 992631 GAGCAGUUCCGCCAAAUCGAGGAACUCUACUUGACAUUUGCCAAGAAAGCUUCGGCCUUCAAUUCUUGGUUCGAAAAUGCCGAAGAGGAUCUCACAGAUCCUGUUCGCUGCAAUUCGAUC .((.((((((..........))))))))...........((((((((.((....))......))))))))(((((..(((((((.((((((.....)))))).))))..))).))))).. ( -39.80, z-score = -2.88, R) >dp4.chrXR_group8 522749 120 - 9212921 GAGCAGUUCCGCCAAAUCGAGGAACUCUACUUGACAUUUGCCAAGAAAGCUUCGGCCUUCAAUUCUUGGUUCGAAAAUGCCGAAGAGGAUCUCACAGAUCCUGUUCGCUGCAAUUCGAUC .((.((((((..........))))))))...........((((((((.((....))......))))))))(((((..(((((((.((((((.....)))))).))))..))).))))).. ( -39.80, z-score = -2.88, R) >droAna3.scaffold_13337 22237841 120 + 23293914 GAGCAGUUCCGCCAGAUCGAGGAACUCUACUUGACAUUUGCCAAGAAAGCUUCGGCCUUCAAUUCUUGGUUCGAGAAUGCCGAAGAGGAUCUCACAGAUCCUGUUCGCUGCAACUCGAUC (.((......))).(((((((........(((((.....((((((((.((....))......)))))))))))))..(((((((.((((((.....)))))).))))..))).))))))) ( -42.90, z-score = -2.79, R) >droEre2.scaffold_4784 1766100 120 - 25762168 GAGCAGUUCCGCCAGAUCGAGGAACUCUACUUGACAUUUGCCAAGAAAGCUUCGGCCUUCAAUUCUUGGUUCGAAAAUGCCGAGGAGGAUCUUACAGAUCCUGUUCGCUGCAAUUCGAUC .((.((((((..........))))))))...........((((((((.((....))......))))))))(((((..(((((((.((((((.....)))))).))))..))).))))).. ( -39.50, z-score = -2.19, R) >droYak2.chr3L 1733798 120 - 24197627 GAGCAGUUCCGCCAGAUCGAGGAACUCUACUUGACAUUUGCCAAGAAAGCUUCGGCCUUCAAUUCUUGGUUCGAAAAUGCCGAGGAGGAUCUCACAGAUCCUGUUCGCUGCAAUUCGAUC .((.((((((..........))))))))...........((((((((.((....))......))))))))(((((..(((((((.((((((.....)))))).))))..))).))))).. ( -39.50, z-score = -1.97, R) >droSec1.super_2 1807598 120 - 7591821 GAGCAGUUCCGCCAGAUCGAGGAACUCUACUUGACAUUUGCCAAGAAAGCUUCGGCCUUCAAUUCUUGGUUCGAAAAUGCCGAGGAGGAUCUCACAGAUCCUGUUCGCUGCAAUUCGAUC .((.((((((..........))))))))...........((((((((.((....))......))))))))(((((..(((((((.((((((.....)))))).))))..))).))))).. ( -39.50, z-score = -1.97, R) >droSim1.chr3L 1337818 120 - 22553184 GAGCAGUUCCGCCAGAUCGAGGAACUCUACUUGACAUUUGCCAAGAAAGCUUCGGCCUUCAAUUCUUGGUUCGAAAAUGCCGAGGAGGAUCUCACAGAUCCUGUUCGCUGCAAUUCGAUC .((.((((((..........))))))))...........((((((((.((....))......))))))))(((((..(((((((.((((((.....)))))).))))..))).))))).. ( -39.50, z-score = -1.97, R) >apiMel3.Group3 1563021 120 + 12341916 GAUCAAUUCAGACAAAUUGAAGAAUUGUAUUUAACAUUUGCCAAGAAAGCAUCUGCUUUCAAUUCAUGGUUUGAAAAUGCAGAAGAAGAUUUAACAGAUCCUGUACGAUGUAAUAGUAUU .(((..(((((((....((((....(((.....)))........((((((....))))))..))))..)))))))..(((((.....((((.....))))))))).)))........... ( -22.20, z-score = -0.51, R) >triCas2.ChLG2 749421 120 - 12900155 GACCAAUUCCGACAAAUCGAAGAUCUUUUCCUCACUUUUGCCAAGAGAGCGUCUGCGUUCAAUUCUUGGUUUGAAAAUGCUGAGGAGGAUUUGACCGAUCCAGUGCGAUGCAAUUCCAUU .........((.((.((((.((((((..(((((((((((((((((((((((....)))))...)))))))...)))).).))))))))))))...))))....))))............. ( -30.80, z-score = -0.69, R) >consensus GAGCAGUUCCGCCAAAUCGAGGAACUCUACUUGACAUUUGCCAAGAAAGCUUCGGCCUUCAAUUCUUGGUUCGAAAAUGCCGAGGAGGAUCUCACAGAUCCUGUUCGCUGCAAUUCGAUC ................(((((........))))).....((((((((.((....))......))))))))(((((..(((((.(.((((((.....)))))).).))..))).))))).. (-26.32 = -26.39 + 0.08)

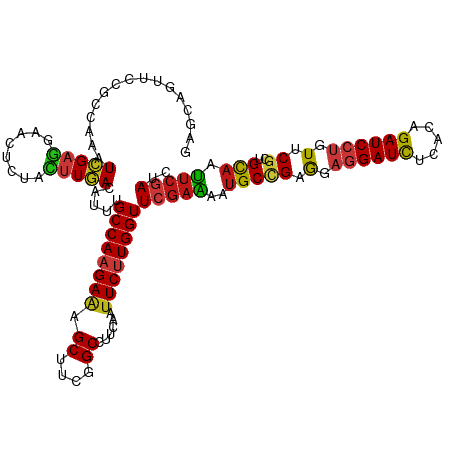
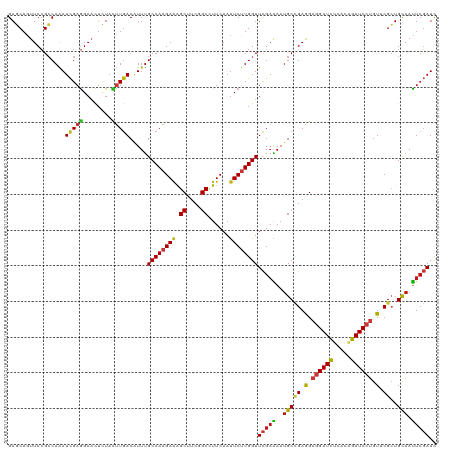
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:56:33 2011