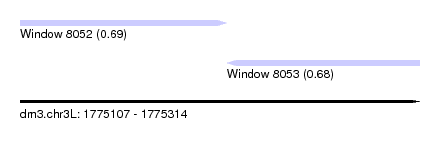
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 1,775,107 – 1,775,314 |
| Length | 207 |
| Max. P | 0.692418 |

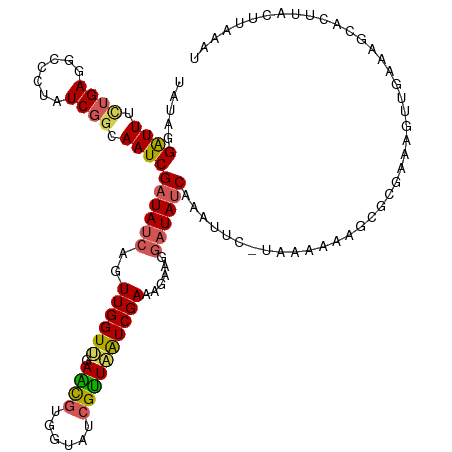
| Location | 1,775,107 – 1,775,214 |
|---|---|
| Length | 107 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 74.29 |
| Shannon entropy | 0.47486 |
| G+C content | 0.35923 |
| Mean single sequence MFE | -27.02 |
| Consensus MFE | -11.88 |
| Energy contribution | -11.96 |
| Covariance contribution | 0.08 |
| Combinations/Pair | 1.29 |
| Mean z-score | -1.87 |
| Structure conservation index | 0.44 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.43 |
| SVM RNA-class probability | 0.692418 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 1775107 107 + 24543557 UAUAGGGUUUCAGAGGCCCUAUCGGCAAUCGAUAUCAGUUGGUUGAACUUGGUAUCGUUAGUCGAUAGGAGGAUAUCAAUUUUGUAUAAAAGCGCGUUUAUUUAAAC------------- ...........(((.(((((((((((...(((((((((((.....))).))))))))...)))))))))..............((......)))).)))........------------- ( -28.00, z-score = -2.35, R) >droEre2.scaffold_4784 1759785 111 + 25762168 UAUAGGAUUUGUGAGGCCCUAUCGGCAAUCGAUAUCAGUUGGUAGAGUGUAGUAUCGUUCAUCGA--------UAACAAAUUC-UAUCAAAGCGCGAAAGUUGGGAUAACUUAUUUAAAU ..........(((((.((((((((.....)))))....((((((((((....(((((.....)))--------))....))))-))))))...((....)).)))....)))))...... ( -28.10, z-score = -2.13, R) >droYak2.chr3L 1727564 112 + 24197627 UAUGGGAUUUUUGA-----UAUCGGCAAUCGAUAUCAGUUGGUAUAGUGUGGUAUCGCUUAUCGAUAUCUUGAUAUCAAAUUU-UUACAAAGCGCGAAAA--AAUGUUCUUAAACUAAAU ..(((((...((((-----(((((.....)))))))))(((((((.....(((((((.....)))))))...)))))))....-................--.....)))))........ ( -23.20, z-score = -1.27, R) >droSec1.super_2 1801330 119 + 7591821 UAUAGGAUUUCUGAGGGCCUAUCGGCAAUCGAUAUCAGUUGGUUGAACGUGGUAUCGUUAAUCGAAAGAAGGAUAUCAAAUUC-UAAAAAAGCGCGAAAGUUGGAAGCAUUUACUUAAAU ..((((((((..((..(((....)))..))((((((..((((((((.((......))))))))))......))))))))))))-)).....((((....)).....))............ ( -27.90, z-score = -1.62, R) >droSim1.chr3L 1331545 119 + 22553184 UAUAGGAUUUCUGAGGGCCUAUCGGCAAUCGAUAUCAGUUGGUUGAACGUGGUAUCGUUAAUCGAAAAAAGGAUAUCAAAUUC-UAAAAAAGCGCGAAAGUUGAAAGCACUUACUUAAAU ..((((((((..((..(((....)))..))((((((..((((((((.((......))))))))))......))))))))))))-)).....((((....)).....))............ ( -27.90, z-score = -1.98, R) >consensus UAUAGGAUUUCUGAGGCCCUAUCGGCAAUCGAUAUCAGUUGGUUGAACGUGGUAUCGUUAAUCGAAAGAAGGAUAUCAAAUUC_UAAAAAAGCGCGAAAGUUGAAAGCACUUACUUAAAU .....((((.((((.......)))).))))((((((..((((((.((((......))))))))))......))))))........................................... (-11.88 = -11.96 + 0.08)

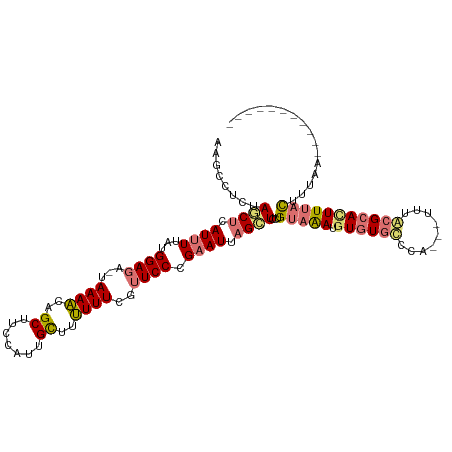
| Location | 1,775,214 – 1,775,314 |
|---|---|
| Length | 100 |
| Sequences | 4 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 71.19 |
| Shannon entropy | 0.44080 |
| G+C content | 0.35004 |
| Mean single sequence MFE | -20.90 |
| Consensus MFE | -10.01 |
| Energy contribution | -10.82 |
| Covariance contribution | 0.81 |
| Combinations/Pair | 1.24 |
| Mean z-score | -1.84 |
| Structure conservation index | 0.48 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.40 |
| SVM RNA-class probability | 0.677998 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 1775214 100 - 24543557 AAGCCUAUAACUUAUUUUAUGGAAA--AAAAUCGCGUUCAUUGCUUUUUU-GCUCCUGAAUAAGGUGGUGCAAAUGUAUGCCCA---UUUACGCAUUUUACUUUAA----------- ..((......(((((((...(((..--((((..(((.....)))..))))-..))).)))))))((((.(((......))))))---)....))............----------- ( -22.00, z-score = -1.84, R) >droEre2.scaffold_4784 1759896 114 - 25762168 GAGCUUCCAUCUCAUGUUAUGGAGAUUAAAGCUGCUUCCUUUGCUCCUUUUGUUCCCGAAUCUGGUCCUGUAGAUUUGUAUCCA---UUUACGCACUUCACUUGCCAUGUAAAUUAA .(((((..(((((........)))))..)))))((.......))............((((((((......)))))))).....(---((((((((.......)))...))))))... ( -20.20, z-score = -0.64, R) >droSec1.super_2 1801449 105 - 7591821 AAUUCUCUAGCUCAUUUUAUGGAGA-UAAAACAGCUUCCAUUGUUUUUUUCGUUCCCGAAUUAGCUCCUGUAAAUGUGUGCUCAUUAUUUGCGCACUUUAUUUUUA----------- ........((((.((((...((((.-.(((((((......))))))).....)))).)))).))))...(((((.((((((.........))))))))))).....----------- ( -20.70, z-score = -2.49, R) >droSim1.chr3L 1331664 105 - 22553184 AAUUCUCUAGCUCAUUUUAUGGAGA-UAAAACAGCUUCCAUUGUUUUUUUCGUUCCCGAAUUAGCUCCUGUAAAUGUGUGCUCAUUAUUUGCGCACUUUAUUUUAA----------- ........((((.((((...((((.-.(((((((......))))))).....)))).)))).))))...(((((.((((((.........))))))))))).....----------- ( -20.70, z-score = -2.41, R) >consensus AAGCCUCUAGCUCAUUUUAUGGAGA_UAAAACAGCUUCCAUUGCUUUUUUCGUUCCCGAAUUAGCUCCUGUAAAUGUGUGCCCA___UUUACGCACUUUACUUUAA___________ ........((((.((((...((((...((((..((.......))..))))..)))).)))).))))...(((((.((((((.........)))))))))))................ (-10.01 = -10.82 + 0.81)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:56:29 2011