
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 1,749,454 – 1,749,568 |
| Length | 114 |
| Max. P | 0.864190 |

| Location | 1,749,454 – 1,749,568 |
|---|---|
| Length | 114 |
| Sequences | 14 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 72.88 |
| Shannon entropy | 0.61507 |
| G+C content | 0.57386 |
| Mean single sequence MFE | -39.34 |
| Consensus MFE | -18.95 |
| Energy contribution | -18.75 |
| Covariance contribution | -0.20 |
| Combinations/Pair | 1.77 |
| Mean z-score | -1.27 |
| Structure conservation index | 0.48 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.97 |
| SVM RNA-class probability | 0.864190 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
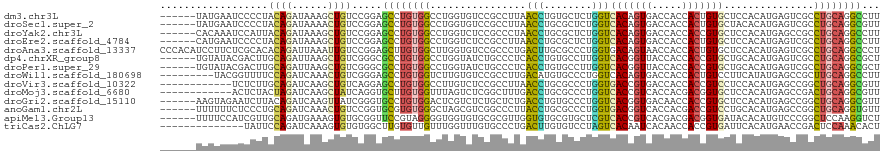
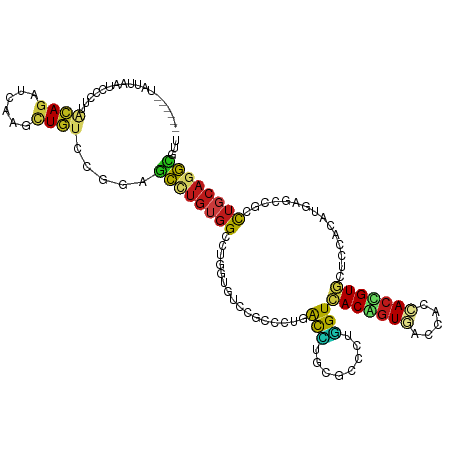
>dm3.chr3L 1749454 114 + 24543557 ------UAUGAAUCCCCUACAGAUAAAGCUGUCCGGAGCCUGUGGCCUGGUGUCCGCCUUAACCUGUGCUCUGGUCACAGUGACCACCACUGUGCUCCACAUGAGUCGCCUGCAGGCCUU ------......(((...((((......))))..)))(((((((((..(((....))).........(((((((.(((((((.....)))))))..)))...)))).))).))))))... ( -39.30, z-score = -1.89, R) >droSec1.super_2 1775340 114 + 7591821 ------UAUGAAUCCCCUACAGAUAAAACUGUCCGGAGCCUGUGGCCUGGUGUCCGCCUUAACCUGCGCUCUGGUCACAGUGACCACCACUGUGCUACACAUGAGUCGCCUGCAGGCGUU ------......(((...((((......))))..)))(((((..(...(((....))).......(((((((((((((((((.....)))))))..)).)).))).))))..)))))... ( -39.50, z-score = -2.05, R) >droYak2.chr3L 1701831 114 + 24197627 ------CACAAAUCCAUUACAGAUAAAGCUGUCCGGAGCCUGUGGCCUGGUCUCCGCCCUAACCUGCGCUCUGGUCACAGUGACCACCACCGUGCUCCACAUGAGCCGCCUGCAGGCCUU ------......(((...((((......))))..)))(((((((((.((((...(((........)))....((((.....))))))))..(.((((.....)))))))).))))))... ( -35.90, z-score = -1.14, R) >droEre2.scaffold_4784 1734087 114 + 25762168 ------CAUGAAUCCCCUACAGAUAAAGCUGUCCGGAGCCUGUGGCCUGGUCUCCGCCUUAACCUGCGCUCUGGUCACAGUGACCACCACUGUGCUCCACAUGAGUCGCCUGCAGGCCUU ------......(((...((((......))))..)))(((((..(...(((....))).......(((((((((.(((((((.....)))))))..)))...))).))))..)))))... ( -39.30, z-score = -1.92, R) >droAna3.scaffold_13337 22206113 120 - 23293914 CCCACAUCCUUCUCGCACACAGAUUAAAUUGUCCGGAGCUUGUGGCUUGGUGUCCGCCCUGACUUGCGCCCUGGUGACAGUAACCACCACUGUGCUCCACAUGAGUCGCCUGCAGGCCCU .....................(((......))).((.(((((((((..(((....)))..(((((((((..(((((........)))))..)))).......)))))))).)))))))). ( -34.31, z-score = 0.02, R) >dp4.chrXR_group8 5111496 114 - 9212921 ------UGUAUACGACUUGCAGAUUAAGCUGUCGGGCGCCUGUGGCCUGGUAUCUGCCCUCACCUGUGCCUUGGUCACGGUUACCACCACCGUGCUGCACAUGAGUCGCCUGCAGGCGCU ------......((((..((.......)).))))((((((((((((..(((....)))((((..(((((...((.((((((.......)))))))))))))))))..))).))))))))) ( -48.00, z-score = -2.06, R) >droPer1.super_29 765143 114 - 1099123 ------UGUAUACGACUUGCAGAUUAAGCUGUCGGGCGCCUGUGGCCUGGUAUCUGCCCUCACCUGUGCCUUGGUCACGGUUACCACCACCGUGCUGCACAUGAGUCGCCUGCAGGCGCU ------......((((..((.......)).))))((((((((((((..(((....)))((((..(((((...((.((((((.......)))))))))))))))))..))).))))))))) ( -48.00, z-score = -2.06, R) >droWil1.scaffold_180698 1687518 111 - 11422946 ---------UACGGUUUUCCAGAUCAAACUGUCGGGAGCCUGUGGUCUUGUGUCCGCCUUGACAUGUGCCCUGGUCACAGUGACCACCACUGUCCUUCAUAUGAGCCGCUUGCAGGCCUU ---------...((((((((.(((......))).)))((..((((.((..(((.......((((.(((...(((((.....))))).)))))))....)))..))))))..))))))).. ( -33.80, z-score = -0.19, R) >droVir3.scaffold_10322 324655 108 - 456481 ------------UCUCUUGCAGAUCAAGCUGUCAGGAGCCUGUGGCCUUGUCUCCGCCUUAACCUGCGCCCUGGUGACCGUGACCACCACCGUCCUCCACAUGAGCCGGCUGCAGGCGUU ------------..((((((((......))).)))))(((((..(((..(.((((((........)))...(((.(((.(((.....))).)))..)))...)))).)))..)))))... ( -39.70, z-score = -1.53, R) >droMoj3.scaffold_6680 22587729 108 - 24764193 ------------ACUCUACUAGAUCAAGCUAUCAGGUGCUUGUGGUUUAGUCUCGGCUUUGACCUGCGCCCUGGUCACCGUCACCACGACGGUGCUCCACAUGAGCCGACUGCAGGCGUU ------------............(((((........))))).((((.(((....)))..)))).(((((..((((((((((.....))))))((((.....)))).))))...))))). ( -36.90, z-score = -0.93, R) >droGri2.scaffold_15110 375619 114 + 24565398 ------AAGUAGAAUCUUACAGAUCAAGUUAUCGGGUGCCUGUGGACUCGUCUCUGCUCUGACCUGUGCCCUGGUCACGGUGACAACCACCGUGCUCCACAUGAGCCGGCUGCAGGCGUU ------..((((....)))).................(((((..(....(((.(((...(((((........)))))))).))).....(((.((((.....))))))))..)))))... ( -36.90, z-score = -0.07, R) >anoGam1.chr2L 17472119 114 + 48795086 ------UUUUUUCUCCCUGCAGAUCAAACUGUCCGGUGCGUGUGGGCUAGCGUCGGCCCUUACCUGCGCCCUGGUGACGGUCACCACGACCGUCCUGCACAUGAGCCGGCUGCAGGUGUU ------.........(((((((..........(((((.((((((.((((((((.((......)).))))..))))(((((((.....)))))))...)))))).)))))))))))).... ( -48.30, z-score = -2.31, R) >apiMel3.Group13 1306447 114 + 8929068 ------UUUUCCAUCGUUGCAGAUGAAAGUGUGCGGUUCCGUAGGGGUGGUGUGCGCGUUGGUGUGCGUGCUCGUCACCGUCACGACGACGGUGAUACACAUGUCCCGGCUCCAAGGUCU ------....((......(((.((....)).)))((..((...(((..(((((((((((......))))))..(((((((((.....))))))))))))).)..)))))..))..))... ( -45.60, z-score = -1.37, R) >triCas2.ChLG7 4599775 106 + 17478683 --------------UAUUCCAGAUCAAAGUGUGUGGCUUGUGUUGUUUGGUUUGUGCCCUGACUUGUGUCCUAGUCACAAUCACAACCACCGUGAUUCACAUGAACCGACUCCAAACACU --------------....(((.((......)).)))...(((((..((((((..((...(((((........))))).((((((.......))))))..))..)))))).....))))). ( -25.20, z-score = -0.26, R) >consensus ______UAUUAAUCCCUUACAGAUCAAGCUGUCCGGAGCCUGUGGCCUGGUGUCCGCCCUGACCUGCGCCCUGGUCACAGUGACCACCACCGUGCUCCACAUGAGCCGCCUGCAGGCGUU ..................((((......)))).....((((((((................(((........)))(((((((.....)))))))...............))))))))... (-18.95 = -18.75 + -0.20)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:56:25 2011