| Sequence ID | dm3.chr3L |
|---|---|
| Location | 1,739,849 – 1,739,941 |
| Length | 92 |
| Max. P | 0.963494 |

| Location | 1,739,849 – 1,739,941 |
|---|---|
| Length | 92 |
| Sequences | 6 |
| Columns | 101 |
| Reading direction | forward |
| Mean pairwise identity | 80.00 |
| Shannon entropy | 0.36646 |
| G+C content | 0.42462 |
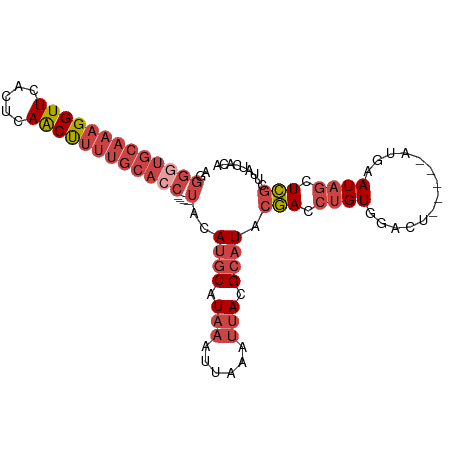
| Mean single sequence MFE | -24.87 |
| Consensus MFE | -16.29 |
| Energy contribution | -17.67 |
| Covariance contribution | 1.38 |
| Combinations/Pair | 1.20 |
| Mean z-score | -1.94 |
| Structure conservation index | 0.66 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.20 |
| SVM RNA-class probability | 0.907834 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
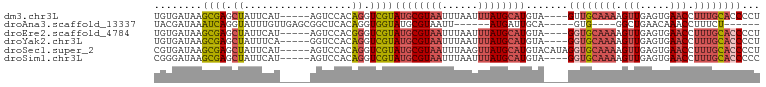
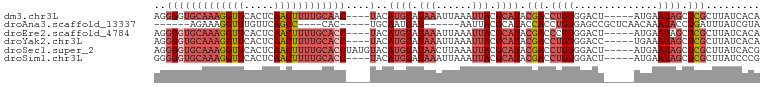
>dm3.chr3L 1739849 92 + 24543557 UGUGAUAAGCGAGCUAUUCAU-----AGUCCACAGGUCGUAUGCGUAAUUUAAUUUAUGCAUGUA----GUUGCAAAAGUUGAGUGAACCUUUGCACCCCU ...(((..(.(..(((....)-----))..).)..))).(((((((((......)))))))))..----..((((((.(((.....))).))))))..... ( -20.30, z-score = -0.94, R) >droAna3.scaffold_13337 22193876 80 - 23293914 UACGAUAAAUCAGGUAUUUGUUGAGCGGCUCACAGGUGGUAUGCGUAAUU------AUGAUUGCA-----GUG----GGCUGAACAAACCUUUCU------ ...........((((..(((((...((((((((..((.....))(((((.------...))))).-----)))----))))))))))))))....------ ( -19.90, z-score = -0.82, R) >droEre2.scaffold_4784 1723326 92 + 25762168 UGUGAUAAGCGAGCUAUUCAU-----AGUCCACGGGUCGUAUGCGUAAUUUAAUUUAUGCAUGUA----GGUGCAAAAGUUGAGUGAACCUUUGCACCCCU ...(((..(.(..(((....)-----))..).)..))).(((((((((......)))))))))..----((((((((.(((.....))).))))))))... ( -27.60, z-score = -2.57, R) >droYak2.chr3L 1691933 92 + 24197627 UGUGAUAAGCGAGCUAUUUCA-----GGUCCACAGGUCGUAUGCGUAAUUUAAUUUAUGCAUGUA----GGUGCAAAAGUUGAGUGAACCUUUGCACCCCU ...(((..(.(..((......-----))..).)..))).(((((((((......)))))))))..----((((((((.(((.....))).))))))))... ( -25.70, z-score = -1.93, R) >droSec1.super_2 1765975 96 + 7591821 CGUGAUAAGCGAGCUAUUCAU-----AGUCCACAGGUCGUAUGCGUAAUUUAAGUUAUGCAUGUACAUAGGUGCAAAAGUUGAGUGAACCUUUGCACCCCU ...(((..(.(..(((....)-----))..).)..))).((((((((((....))))))))))......((((((((.(((.....))).))))))))... ( -27.30, z-score = -2.47, R) >droSim1.chr3L 1294525 92 + 22553184 CGGGAUAAGCGAGCUAUUCAU-----AGUCCACAGGUCGUAUGCGUAAUUUAAUUUAUGCAUGUA----GGUGCAAAAGUUGAGUGAACCUUUGCACCCCC .(((....((((.((......-----.......)).))))((((((((......))))))))...----((((((((.(((.....))).))))))))))) ( -28.42, z-score = -2.88, R) >consensus UGUGAUAAGCGAGCUAUUCAU_____AGUCCACAGGUCGUAUGCGUAAUUUAAUUUAUGCAUGUA____GGUGCAAAAGUUGAGUGAACCUUUGCACCCCU ........((((.((..................)).))))((((((((......)))))))).......((((((((.(((.....))).))))))))... (-16.29 = -17.67 + 1.38)
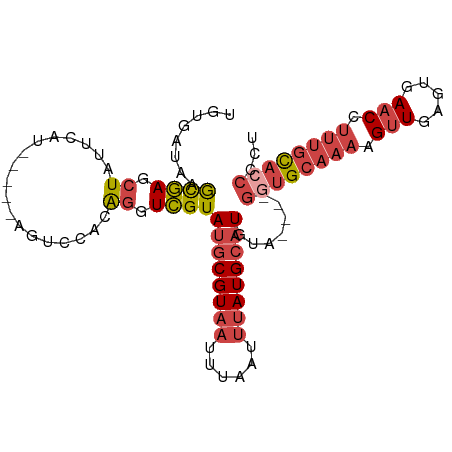
| Location | 1,739,849 – 1,739,941 |
|---|---|
| Length | 92 |
| Sequences | 6 |
| Columns | 101 |
| Reading direction | reverse |
| Mean pairwise identity | 80.00 |
| Shannon entropy | 0.36646 |
| G+C content | 0.42462 |
| Mean single sequence MFE | -23.29 |
| Consensus MFE | -13.57 |
| Energy contribution | -15.56 |
| Covariance contribution | 1.99 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.51 |
| Structure conservation index | 0.58 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.72 |
| SVM RNA-class probability | 0.963494 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 1739849 92 - 24543557 AGGGGUGCAAAGGUUCACUCAACUUUUGCAAC----UACAUGCAUAAAUUAAAUUACGCAUACGACCUGUGGACU-----AUGAAUAGCUCGCUUAUCACA (((((((((((((((.....))))))))).))----)..((((.(((......))).))))....)))(((..((-----(....)))..)))........ ( -21.40, z-score = -1.98, R) >droAna3.scaffold_13337 22193876 80 + 23293914 ------AGAAAGGUUUGUUCAGCC----CAC-----UGCAAUCAU------AAUUACGCAUACCACCUGUGAGCCGCUCAACAAAUACCUGAUUUAUCGUA ------....(((((((((.(((.----...-----.((..((((------(...............))))))).))).)))))..))))........... ( -10.76, z-score = 0.41, R) >droEre2.scaffold_4784 1723326 92 - 25762168 AGGGGUGCAAAGGUUCACUCAACUUUUGCACC----UACAUGCAUAAAUUAAAUUACGCAUACGACCCGUGGACU-----AUGAAUAGCUCGCUUAUCACA .((((((((((((((.....))))))))))))----...((((.(((......))).)))).....))(((..((-----(....)))..)))........ ( -26.70, z-score = -3.57, R) >droYak2.chr3L 1691933 92 - 24197627 AGGGGUGCAAAGGUUCACUCAACUUUUGCACC----UACAUGCAUAAAUUAAAUUACGCAUACGACCUGUGGACC-----UGAAAUAGCUCGCUUAUCACA ..(((((((((((((.....))))))))))))----)..((((.(((......))).)))).(((.((((.....-----....)))).)))......... ( -25.10, z-score = -2.86, R) >droSec1.super_2 1765975 96 - 7591821 AGGGGUGCAAAGGUUCACUCAACUUUUGCACCUAUGUACAUGCAUAACUUAAAUUACGCAUACGACCUGUGGACU-----AUGAAUAGCUCGCUUAUCACG (((((((((((((((.....))))))))))))..((((..(((.(((......))).))))))).)))(((..((-----(....)))..)))........ ( -26.90, z-score = -3.09, R) >droSim1.chr3L 1294525 92 - 22553184 GGGGGUGCAAAGGUUCACUCAACUUUUGCACC----UACAUGCAUAAAUUAAAUUACGCAUACGACCUGUGGACU-----AUGAAUAGCUCGCUUAUCCCG (((((((((((((((.....))))))))))))----...((((.(((......))).)))).......(((..((-----(....)))..)))....))). ( -28.90, z-score = -3.98, R) >consensus AGGGGUGCAAAGGUUCACUCAACUUUUGCACC____UACAUGCAUAAAUUAAAUUACGCAUACGACCUGUGGACU_____AUGAAUAGCUCGCUUAUCACA ...((((((((((((.....)))))))))))).......((((.(((......))).)))).(((.((((..............)))).)))......... (-13.57 = -15.56 + 1.99)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:56:23 2011