| Sequence ID | dm3.chr2L |
|---|---|
| Location | 4,138,880 – 4,138,972 |
| Length | 92 |
| Max. P | 0.978191 |

| Location | 4,138,880 – 4,138,972 |
|---|---|
| Length | 92 |
| Sequences | 8 |
| Columns | 92 |
| Reading direction | reverse |
| Mean pairwise identity | 78.59 |
| Shannon entropy | 0.45830 |
| G+C content | 0.53229 |
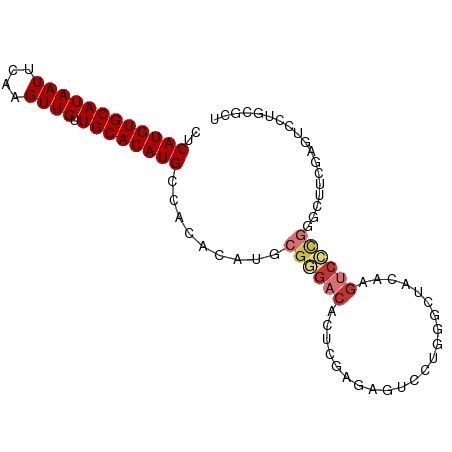
| Mean single sequence MFE | -31.96 |
| Consensus MFE | -15.59 |
| Energy contribution | -16.28 |
| Covariance contribution | 0.69 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.54 |
| Structure conservation index | 0.49 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.99 |
| SVM RNA-class probability | 0.978191 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
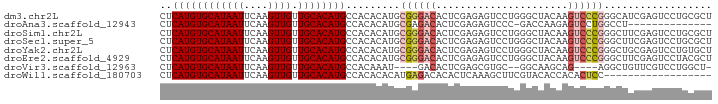
>dm3.chr2L 4138880 92 - 23011544 CUCAUGUGCAUAAUUCAAGUUGUUGCACAUGCCACACAUGCGGGACACUCGAGAGUCCUGGGCUACAAGUCCCGGGCAUCGAGUCCUGCGCU ..((((((((((((....)))).))))))))........(((((((..((((..((((.((((.....)))).)))).)))))))))))... ( -40.40, z-score = -3.90, R) >droAna3.scaffold_12943 708827 77 + 5039921 CUCAUGUGCAUAAUUCAAGUUGUUGCACAUGCCACACAUGCGAGACACUCGAGAGUCCC-GACCAAGAGUCCUGGCCU-------------- ..((((((((((((....)))).))))))))........((.((..((((..(.((...-.)))..)))).)).))..-------------- ( -19.50, z-score = -0.76, R) >droSim1.chr2L 4094774 92 - 22036055 CUCAUGUGCAUAAUUCAAGUUGUUGCACAUGCCACACAUGCGGGACACUCGAGAGUCCUGGGCUACAAGUCCCGGGCUUCGAGUCCUGCGCU ..((((((((((((....)))).))))))))........(((((((..((((.(((((.((((.....)))).))))))))))))))))... ( -41.40, z-score = -4.26, R) >droSec1.super_5 2238004 92 - 5866729 CUCAUGUGCAUAAUUCAAGUUGUUGCACAUGCCACACAUGCGGGACACUCGAGAGUCCUGGGCUACAAGUCCCGGGCUUCGAGUCCUGCGCU ..((((((((((((....)))).))))))))........(((((((..((((.(((((.((((.....)))).))))))))))))))))... ( -41.40, z-score = -4.26, R) >droYak2.chr2L 4161071 92 - 22324452 CUCAUGUGCAUAAUUCAAGUUGUUGCACAUGCCACACAUGCGGGACACUCGAGAGUCCUGGGCUACAAGUCCCGGGCUGCGAGUCCUGUGCU ..((((((((((((....)))).))))))))........(((((((..(((..(((((.((((.....)))).))))).))))))))))... ( -38.10, z-score = -2.70, R) >droEre2.scaffold_4929 4202465 92 - 26641161 CUCAUGUGCAUAAUUCAAGUUGUUGCACAUGCCACACAUGCGGGACACUCGAGAGUCCUGGGCUACAAGUCCCGGGCUUCGAGUCCUACGCU ..((((((((((((....)))).))))))))........(((....((((((.(((((.((((.....)))).)))))))))))....))). ( -37.60, z-score = -3.64, R) >droVir3.scaffold_12963 11139537 81 + 20206255 CUCAUGUGCAUAAUUCAAGUUGUUGCACAUGCCACAAAU----GACACUCGAGCGUGC--GGCAAGCAG----AGGCUGUUCGUCCUGGCU- ...(((((((((((....)))).)))))))((((.....----(.....)(..((.((--(((......----..))))).))..))))).- ( -23.10, z-score = 0.35, R) >droWil1.scaffold_180703 2140093 74 - 3946847 CUCAUGUGCAUAAUUCAAGUUGUUGCACAUGCCACACACAUGAGACACACUCAAAGCUUCGUACACCACACUCC------------------ ..((((((((((((....)))).)))))))).........((((.....)))).....................------------------ ( -14.20, z-score = -1.13, R) >consensus CUCAUGUGCAUAAUUCAAGUUGUUGCACAUGCCACACAUGCGGGACACUCGAGAGUCCUGGGCUACAAGUCCCGGGCUUCGAGUCCUGCGCU ..((((((((((((....)))).)))))))).........((((((......................)))))).................. (-15.59 = -16.28 + 0.69)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:15:28 2011