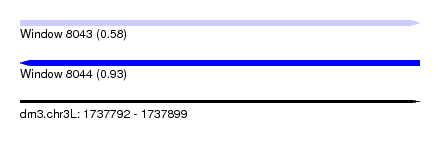
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 1,737,792 – 1,737,899 |
| Length | 107 |
| Max. P | 0.932289 |

| Location | 1,737,792 – 1,737,899 |
|---|---|
| Length | 107 |
| Sequences | 8 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 71.79 |
| Shannon entropy | 0.54894 |
| G+C content | 0.51791 |
| Mean single sequence MFE | -34.39 |
| Consensus MFE | -10.24 |
| Energy contribution | -11.74 |
| Covariance contribution | 1.50 |
| Combinations/Pair | 1.15 |
| Mean z-score | -2.16 |
| Structure conservation index | 0.30 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.18 |
| SVM RNA-class probability | 0.580912 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
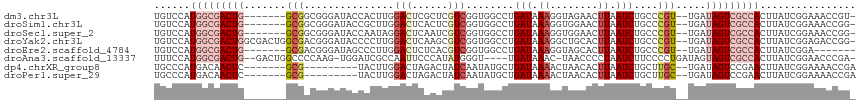
>dm3.chr3L 1737792 107 + 24543557 UGUCCAUGGCGACUG-------GCGGCGGGAUACCACUUGGACUCGCUCGUCGGUGGCCUGAUAAAGGUAGAACUUAAUCUGCCCGU--UGAUAGUCGCCACUUAUCGGAAACCGU- ......(((((((((-------.(((((((...(((((..(((......))))))))...(((.(((......))).)))..)))))--)).)))))))))......(....)...- ( -40.00, z-score = -1.94, R) >droSim1.chr3L 1292530 107 + 22553184 UGUCCAUGGCGACUG-------GCGGCGGGAUACCGCUUGGACUCACUCGUCGGUGGCCUGAUAAAGGUGGAACUUAAUCUGCCCGU--UGAUAGUCGCCACUUAUCGGAAACCGG- ...((.(((((((((-------.(((((((...(((((..(((......))))))))...(((.(((......))).)))..)))))--)).)))))))))......(....).))- ( -40.40, z-score = -1.75, R) >droSec1.super_2 1763994 107 + 7591821 UGUCCAUGGCGACUG-------GCGGCGGGAUACCAAUAGGACUCAAUCGUCGGUGGCCUGAUAAAGGUGGAACUUAAUCUGCCCGU--UGAUAGUCGCCACUUAUCGGAAACCGG- ...((.(((((((((-------.(((((((.((((.....(((......)))))))((((.....)))).............)))))--)).)))))))))......(....).))- ( -37.90, z-score = -1.44, R) >droYak2.chr3L 1689854 114 + 24197627 UGUCCAUGGCGACUGGCGACUGGCGACGGGAUACCCCUUGGACUCAAGCGUCGGUGGCCUGAUAAAGGCUGCACUUAAUCUGCCCGU--UGAUAGUCGCCACUUAUCGGAGACCGG- .(((..((..((.(((((((((.(((((((..........(((......))).(..((((.....))))..)..........)))))--)).))))))))).))..))..)))...- ( -47.70, z-score = -2.87, R) >droEre2.scaffold_4784 1721185 101 + 25762168 UGUCCAUGGCGACUG-------GCGACGGGAUAGCCCUUGGACUCUCACGUCGGUGGCCUGAUAAAGGUAGCACUUAAUCUGCCCGU--UGAUAGUCGCCACUUAUCGGA------- ......(((((((((-------.(((((((...(((..(.(((......))).).)))..(((.(((......))).)))..)))))--)).))))))))).........------- ( -38.90, z-score = -2.07, R) >droAna3.scaffold_13337 22192030 108 - 23293914 UUUCCAUGGCGACUG--GACUGGCCCCAAG-UGGAUCGCCAAUUCCCAUAUGGGU----UGAUAAAC-UAACCCCUAAUCUUCCCCUGAUAGUAGUCGCCACUUAUCGGAACCCGA- ......(((((((((--((.((((.((...-.))...))))..))).....((((----((......-))))))...................))))))))....((((...))))- ( -31.20, z-score = -1.85, R) >dp4.chrXR_group8 5098871 99 - 9212921 UGCCCAUGACAACUC-------GCG---------UACUUGGACUAGACUAUCAAUAUGCUGAUAAAACUAACACUUAAUCUGCUUGC--UGAUAGUCCGAACUUAUCGGAAAACCGA ...(((..((.....-------..)---------)...)))....((((((((.((.((.(((.((........)).))).)).)).--))))))))........((((....)))) ( -19.50, z-score = -2.66, R) >droPer1.super_29 752467 99 - 1099123 UGCCCAUGACAACUC-------GCG---------UACUUGGACUAGACUAUCAAUAUGCUGAUAAAACUAACACUUAAUCUGCUUGC--UGAUAGUCCGAACUUAUCGGAAAACCGA ...(((..((.....-------..)---------)...)))....((((((((.((.((.(((.((........)).))).)).)).--))))))))........((((....)))) ( -19.50, z-score = -2.66, R) >consensus UGUCCAUGGCGACUG_______GCGGCGGGAUACCACUUGGACUCAAUCGUCGGUGGCCUGAUAAAGGUAGCACUUAAUCUGCCCGU__UGAUAGUCGCCACUUAUCGGAAACCGG_ ......(((((((((.......(((...............(((......)))........(((.((........)).)))....))).....)))))))))................ (-10.24 = -11.74 + 1.50)

| Location | 1,737,792 – 1,737,899 |
|---|---|
| Length | 107 |
| Sequences | 8 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 71.79 |
| Shannon entropy | 0.54894 |
| G+C content | 0.51791 |
| Mean single sequence MFE | -33.82 |
| Consensus MFE | -15.70 |
| Energy contribution | -15.06 |
| Covariance contribution | -0.64 |
| Combinations/Pair | 1.46 |
| Mean z-score | -1.88 |
| Structure conservation index | 0.46 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.37 |
| SVM RNA-class probability | 0.932289 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
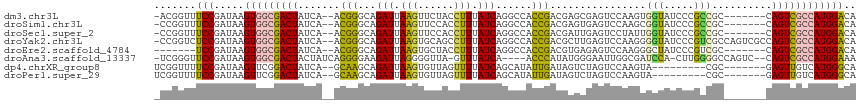
>dm3.chr3L 1737792 107 - 24543557 -ACGGUUUCCGAUAAGUGGCGACUAUCA--ACGGGCAGAUUAAGUUCUACCUUUAUCAGGCCACCGACGAGCGAGUCCAAGUGGUAUCCCGCCGC-------CAGUCGCCAUGGACA -.(((...)))....(((((((((....--.((((..(((.(((......))).)))..(((((.(((......)))...)))))..))))....-------.)))))))))..... ( -34.70, z-score = -1.44, R) >droSim1.chr3L 1292530 107 - 22553184 -CCGGUUUCCGAUAAGUGGCGACUAUCA--ACGGGCAGAUUAAGUUCCACCUUUAUCAGGCCACCGACGAGUGAGUCCAAGCGGUAUCCCGCCGC-------CAGUCGCCAUGGACA -.(((...)))....(((((((((....--..((((.(((.(((......))).)))....(((......))).))))..(((((.....)))))-------.)))))))))..... ( -34.50, z-score = -1.42, R) >droSec1.super_2 1763994 107 - 7591821 -CCGGUUUCCGAUAAGUGGCGACUAUCA--ACGGGCAGAUUAAGUUCCACCUUUAUCAGGCCACCGACGAUUGAGUCCUAUUGGUAUCCCGCCGC-------CAGUCGCCAUGGACA -.(((...)))....(((((((((....--.((((..(((.(((......))).)))..((((..(((......)))....))))..))))....-------.)))))))))..... ( -31.30, z-score = -0.74, R) >droYak2.chr3L 1689854 114 - 24197627 -CCGGUCUCCGAUAAGUGGCGACUAUCA--ACGGGCAGAUUAAGUGCAGCCUUUAUCAGGCCACCGACGCUUGAGUCCAAGGGGUAUCCCGUCGCCAGUCGCCAGUCGCCAUGGACA -...((((.((((...((((((((..(.--(((((........((((..((((..((((((.......))))))....)))).))))))))).)..))))))))))))....)))). ( -40.00, z-score = -1.15, R) >droEre2.scaffold_4784 1721185 101 - 25762168 -------UCCGAUAAGUGGCGACUAUCA--ACGGGCAGAUUAAGUGCUACCUUUAUCAGGCCACCGACGUGAGAGUCCAAGGGCUAUCCCGUCGC-------CAGUCGCCAUGGACA -------(((.....(((((((((....--...(((.(((.(((......))).)))..)))..(((((.((.((((....)))).)).))))).-------.)))))))))))).. ( -37.20, z-score = -2.49, R) >droAna3.scaffold_13337 22192030 108 + 23293914 -UCGGGUUCCGAUAAGUGGCGACUACUAUCAGGGGAAGAUUAGGGGUUA-GUUUAUCA----ACCCAUAUGGGAAUUGGCGAUCCA-CUUGGGGCCAGUC--CAGUCGCCAUGGAAA -((((...))))...(((((((((.(((((.......)).)))(((((.-.......)----)))).....(((.(((((..(((.-...))))))))))--))))))))))..... ( -36.50, z-score = -1.18, R) >dp4.chrXR_group8 5098871 99 + 9212921 UCGGUUUUCCGAUAAGUUCGGACUAUCA--GCAAGCAGAUUAAGUGUUAGUUUUAUCAGCAUAUUGAUAGUCUAGUCCAAGUA---------CGC-------GAGUUGUCAUGGGCA ((((....)))).......(((((((((--(...((.(((.(((......))).))).))...)))))))))).(((((.(.(---------(..-------..))...).))))). ( -28.20, z-score = -3.31, R) >droPer1.super_29 752467 99 + 1099123 UCGGUUUUCCGAUAAGUUCGGACUAUCA--GCAAGCAGAUUAAGUGUUAGUUUUAUCAGCAUAUUGAUAGUCUAGUCCAAGUA---------CGC-------GAGUUGUCAUGGGCA ((((....)))).......(((((((((--(...((.(((.(((......))).))).))...)))))))))).(((((.(.(---------(..-------..))...).))))). ( -28.20, z-score = -3.31, R) >consensus _CCGGUUUCCGAUAAGUGGCGACUAUCA__ACGGGCAGAUUAAGUGCUACCUUUAUCAGGCCACCGACGAGCGAGUCCAAGUGGUAUCCCGCCGC_______CAGUCGCCAUGGACA .......(((.....(((((((((.......(((...(((.(((......))).)))......)))................(((.....)))..........)))))))))))).. (-15.70 = -15.06 + -0.64)
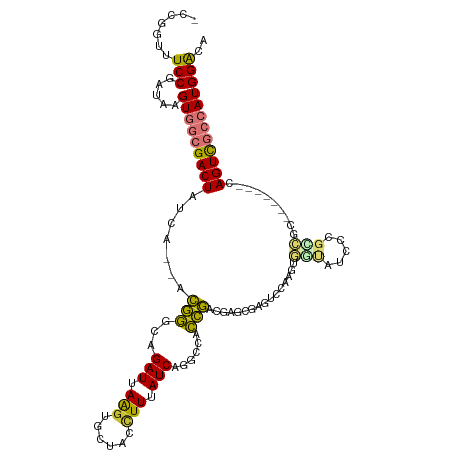

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:56:21 2011