
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 1,710,026 – 1,710,157 |
| Length | 131 |
| Max. P | 0.516157 |

| Location | 1,710,026 – 1,710,157 |
|---|---|
| Length | 131 |
| Sequences | 6 |
| Columns | 163 |
| Reading direction | reverse |
| Mean pairwise identity | 70.10 |
| Shannon entropy | 0.50774 |
| G+C content | 0.58299 |
| Mean single sequence MFE | -51.65 |
| Consensus MFE | -22.37 |
| Energy contribution | -22.68 |
| Covariance contribution | 0.31 |
| Combinations/Pair | 1.39 |
| Mean z-score | -1.57 |
| Structure conservation index | 0.43 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.04 |
| SVM RNA-class probability | 0.516157 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
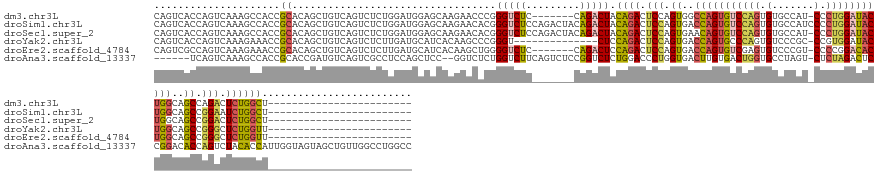
>dm3.chr3L 1710026 131 - 24543557 CAGUCACCAGUCAAAGCCACCGCACAGCUGUCAGUCUCUGGAUGGAGCAAGAACCCGGGUCUC-------CAGACUACAGACUCCAGUGGCCAGUGUCCAGUGUGCCAU-CCCUGGAUACUGGCAGCCAGACUCUGGCU------------------------ .(((((..((((...((.........((((..(((((.(((.((((((..(....)..).)))-------))..))).))))).)))).((((((((((((.(......-).)))))))))))).))..)))).)))))------------------------ ( -49.90, z-score = -1.11, R) >droSim1.chr3L 1268684 139 - 22553184 CAGUCACCAGUCAAAGCCACCGCACAGCUGUCAGUCUCUGGAUGGAGCAAGAACACGGGUCUCCAGACUACAGACUACAGACUCCAGUGACCAGUGUCCAGUGUGCCAUCCCCUGGAUACUGGCAGCCGGAAUCUGGCU------------------------ ..............(((((........((((.(((((.(((.((((((..(....)..).)))))..))).)))))))))..(((.((..(((((((((((.(........))))))))))))..)).)))...)))))------------------------ ( -49.80, z-score = -0.96, R) >droSec1.super_2 1736301 138 - 7591821 CAGUCACCAGUCAAAGCCACCGCACAGCUGUCAGUCUCUGGAUGGAGCAAGAACACGGGUCUCCAGACUACAGACUACAGACUCCAGUGAACAGUGUCCAGUGUGCCAU-CCCUGGAUACUGGCAGCCGGACUCUGGCU------------------------ .(((((..((((...((....)).(.(((((((((.(((((((((.(((...((((((((((..((........))..)))).)).(....).))))....))).))))-))..))).))))))))).))))).)))))------------------------ ( -49.80, z-score = -0.80, R) >droYak2.chr3L 1662002 124 - 24197627 CAGUCACCAGUCAAAGAAACCGCACAGCUGUCAGUCUCUUGAUGCAUCACAAGCCCGGGU--------------CUCCAGACUCCAGUGACCAGUGCCCAGUGUCCCGC-CCGUGGAUACUGGCAGCCGGGCUCUGGUU------------------------ .....(((((................((.(((((....)))))))......(((((((((--------------(....)))............((((.((((((((..-..).))))))))))).)))))))))))).------------------------ ( -42.20, z-score = -0.81, R) >droEre2.scaffold_4784 1693105 131 - 25762168 CAGUCGCCAGUCAAAGAAACCGCACAGCUGUCAGUCUCUUGAUGCAUCACAAGCUGGGGUCUC-------CAGACUCCAGACUCCAGUGACCAGUGUCGAGUGUCCCGU-CCCCGGACACUGGCAGCCGGGCUCUGGUU------------------------ .....(((((....((...(((....(((((((((..((((((((.((((.((((((((((..-------..)))))))).))...))))...)))))))).((((...-....)))))))))))))))).))))))).------------------------ ( -52.30, z-score = -2.08, R) >droAna3.scaffold_13337 22164414 154 + 23293914 ------UCAGUCAAAGCCACCGCACCGAUGUCAGUCGCCUCCAGCUCC--GGUCUCUGGUCUUCAGUCUCCGGUCUCUGGACCCUGGUGACUUGUGACUGGUGCCUAGU-CUCUAGACUCCGGACACCAGUCUACACCAUUGGUAGUAGCUGUUGGCCUGGCC ------...((((..(((((.(((((((((..((((((((((((..((--((...(((.....)))...))))...)))))....)))))))(((((((((((((.(((-(....))))..)).)))))))).))).)))))))....)).).)))).)))). ( -65.90, z-score = -3.67, R) >consensus CAGUCACCAGUCAAAGCCACCGCACAGCUGUCAGUCUCUGGAUGCAGCAAGAACACGGGUCUC_______CAGACUACAGACUCCAGUGACCAGUGUCCAGUGUCCCAU_CCCUGGAUACUGGCAGCCGGACUCUGGCU________________________ .....................((..................................(((((.........))))).((((.(((.((..(((((((((((.(.......).)))))))))))..)).))).))))))......................... (-22.37 = -22.68 + 0.31)


Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:56:17 2011