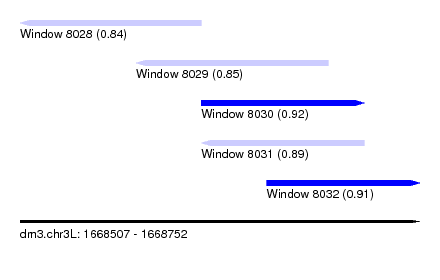
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 1,668,507 – 1,668,752 |
| Length | 245 |
| Max. P | 0.920298 |

| Location | 1,668,507 – 1,668,618 |
|---|---|
| Length | 111 |
| Sequences | 12 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 71.40 |
| Shannon entropy | 0.64364 |
| G+C content | 0.39155 |
| Mean single sequence MFE | -23.89 |
| Consensus MFE | -9.44 |
| Energy contribution | -9.59 |
| Covariance contribution | 0.15 |
| Combinations/Pair | 1.33 |
| Mean z-score | -1.50 |
| Structure conservation index | 0.40 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.86 |
| SVM RNA-class probability | 0.836686 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
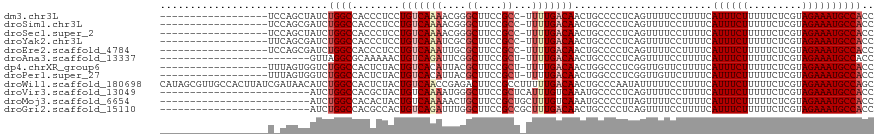
>dm3.chr3L 1668507 111 - 24543557 CAAAAAGCCAACCGAGAAAUCCGGUAAGGAUUUUUCAUUCAAAAAUAUGCGAAAAAUGCGUGAAUCGAUCAGCAAAAGGGAGACGACAGCUCAGCCCACCAGAAGGCAAUC-- ......(((....(((((((((.....))))))))).(((................(((.(((.....))))))...(((.(((....).))..)))....))))))....-- ( -26.00, z-score = -2.02, R) >droSim1.chr3L 1229523 111 - 22553184 CAAAAAGCCAACCGAGAAAUCCGGUAAGGGUUUUUCAUUCAAAAAUAUGCGAAAAAUGCGUGAAUCGAUCAGCAAAAGGAAGGCGACAGCUCAGCCCAUCAGAGGGCAAUC-- ......(((..(((((((((((.....)))))))))....................(((.(((.....))))))...))..))).........((((......))))....-- ( -31.20, z-score = -2.92, R) >droSec1.super_2 1694958 111 - 7591821 CAAAAAGCCAACCGAGAAAUCCGGUAAGGGUUUUUCAUUCAAAAAUAUGCGAAAAAUGCGUGAAUCGAUCAGCAAAAUGGAGGCGACAGCUCAGCCCAUCAGAGGGCAAUC-- ......(((..(((((((((((.....)))))))))....................(((.(((.....))))))....)).))).........((((......))))....-- ( -29.00, z-score = -1.92, R) >droYak2.chr3L 1619264 111 - 24197627 CAAAAAGCCAACCGAGAAAUCCGGUAAGGGUUUUUCAUUCAAAAAUAAGCGAAAAAUGCGUGAAUCGAUCAGCAAAAGCGAGGCGACAGCACGGCCCACCAGCGGGCAAUC-- ......(((....(((((((((.....)))))))))............((......(((.(((.....))))))...))..))).........((((......))))....-- ( -29.40, z-score = -1.62, R) >droEre2.scaffold_4784 1648006 111 - 25762168 CAAAAAGCCAACCGAGAAAUCCGGUAAGGGUUUUUCAUUCAAAAAUAGGCGAAAAAUUCGUGAAUCGAUCAGCAAAAGCGAGGCGACAGCAUGGCCCACCAGCAGGCCAUC-- ......(((....(((((((((.....)))))))))..((........((((.....))))..........((....)))))))......((((((........)))))).-- ( -29.40, z-score = -2.01, R) >droAna3.scaffold_13337 22124536 104 + 23293914 CAAAAAGCCAACCGAGAAAUCCGGUAAGGGUUUUUCAUACAAAAACUGCCCAAAAUUAAGUGAAUUGAUCAGCAAAUGCAACGCGAACAAAAGGGGAAUCCGCU--------- .....(((..(((((....).))))..((((((..(...(((..(((...........)))...)))....((....)).............)..)))))))))--------- ( -20.00, z-score = -0.37, R) >dp4.chrXR_group6 7815924 112 - 13314419 CAAAAAGCCAACUGAGAAAUCCGGUAAGGAUUUUUCAUAUAAAAAUCAGCUAAAAAUGUGUGGAUUUAUCACCAAAAGCAAUGCUAAAGCAUGGAUAAUAAUCAGAAAUAAA- .......((...((((((((((.....))))))))))...................(((.(((........)))...)))((((....))))))..................- ( -20.50, z-score = -1.81, R) >droPer1.super_27 562484 112 + 1201981 CAAAAAGCCAACUGAGAAAUCCGGUAAGGAUUUUUCAUAUAAAAAUCAGCUAAAAAUGUGUGGAUUUAUCACCAAAAGCAAUGCUAAAGCAUGGAUAAUAAUCAGAAAUAAA- .......((...((((((((((.....))))))))))...................(((.(((........)))...)))((((....))))))..................- ( -20.50, z-score = -1.81, R) >droWil1.scaffold_180698 874531 113 - 11422946 UAAAAAGCCAGCCGAAAAAUCCGGUGAGAUAAUUUAAUAGAAUUUUUAACAAAAAAAGUGUGAAAACAAUACCAAAUGAAAGGUUAAAGCAAGUGCUGUGCGUGGAUAUUAAC ......(((((((.........((((.((.(((((....))))).))...........(((....))).)))).........((....))..).)))).))............ ( -14.40, z-score = 0.19, R) >droVir3.scaffold_13049 2286624 113 - 25233164 UAAAAAGCCAACCGAGAAAUCCGGUAAGGAUUUUUCAUAUAAAAAUGUCUAAAAAAUGUGUGGUUUUGUCAUCAGCAGCAAGGUUAAAGCAAGCACAAUGCACGGAUAUAUGU .............(((((((((.....)))))))))........((((((......(((((.((((((..(((........)))))))))..)))))......)))))).... ( -23.20, z-score = -1.34, R) >droMoj3.scaffold_6654 986020 113 + 2564135 UAAAAAGCCAACCGAGAAAUCCGGUAAGGAUUUUUCAUAUAAAAUUAUAUCGAAAAUAUGUGGUUUUGUCACCUAAUUCAAAGUCAAAGCAAGCUGAAUGCAAUGCAGCAUGU ...(((((((...(((((((((.....)))))))))(((((....)))))..........))))))).....................(((.((((.........)))).))) ( -23.40, z-score = -1.81, R) >droGri2.scaffold_15110 3323092 113 - 24565398 UAAAAAGCCAACCGAGAAGUCCGGUAAGCAUUUUUCAUAUAAAAAUGCCAUAAAAAUGUGUGAAUUAAACACCAAACGCUAAACUAAUACAUGGCCAAUGUGCAGCAACGUGU ......((((((((.(....)))))..((((((((.....)))))))).........((((.......))))...................))))..((((......)))).. ( -19.70, z-score = -0.58, R) >consensus CAAAAAGCCAACCGAGAAAUCCGGUAAGGAUUUUUCAUACAAAAAUAUGCGAAAAAUGUGUGAAUUGAUCAGCAAAAGCAAGGCGAAAGCAAGGCCAAUCAGCAGGCAAUC__ .............(((((((((.....)))))))))............................................................................. ( -9.44 = -9.59 + 0.15)
| Location | 1,668,578 – 1,668,696 |
|---|---|
| Length | 118 |
| Sequences | 12 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 90.01 |
| Shannon entropy | 0.22350 |
| G+C content | 0.41907 |
| Mean single sequence MFE | -26.73 |
| Consensus MFE | -19.01 |
| Energy contribution | -19.55 |
| Covariance contribution | 0.54 |
| Combinations/Pair | 1.15 |
| Mean z-score | -2.27 |
| Structure conservation index | 0.71 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.91 |
| SVM RNA-class probability | 0.849630 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 1668578 118 - 24543557 UGUCAAAACGGGCUUCCGCC-UUUUGACAACUGCCCCUCAGUUUUCCUUUUCAUUUCUUUUUCUCGUAGAAAUGCCACCCAAAAAGCCAACCGAGAAAUCCGGUAAGGAUUUUUCAUUC .((((((..((((....)))-)))))))(((((.....)))))........(((((((.........)))))))..................(((((((((.....))))))))).... ( -29.00, z-score = -2.79, R) >droSim1.chr3L 1229594 118 - 22553184 UGUCAAAACGGGCUUCCGCC-UUUUGACAACUGCCCCUCAGUUUUCCUUUUCAUUUCUUUUUCUCGUAGAAAUGCCACCCAAAAAGCCAACCGAGAAAUCCGGUAAGGGUUUUUCAUUC .((((((..((((....)))-)))))))(((((.....)))))........(((((((.........)))))))..................(((((((((.....))))))))).... ( -28.40, z-score = -2.03, R) >droSec1.super_2 1695029 118 - 7591821 UGUCAAAACGGGCUUCCGCC-UUUUGACAACUGCCCCUCAGUUUUCCUUUUCAUUUCUUUUUCUCGUAGAAAUGCCACCCAAAAAGCCAACCGAGAAAUCCGGUAAGGGUUUUUCAUUC .((((((..((((....)))-)))))))(((((.....)))))........(((((((.........)))))))..................(((((((((.....))))))))).... ( -28.40, z-score = -2.03, R) >droYak2.chr3L 1619335 118 - 24197627 UGUCAAAUCGCGCUUCCGCC-UUUUGACAACUGCCCCUCAGUUUUCCUUUUCAUUUCUUUUUCUCGUAGAAAUGCCACCCAAAAAGCCAACCGAGAAAUCCGGUAAGGGUUUUUCAUUC .((((((..(((....))).-.))))))(((((.....)))))........(((((((.........)))))))..................(((((((((.....))))))))).... ( -27.40, z-score = -2.86, R) >droEre2.scaffold_4784 1648077 118 - 25762168 UGUCAAAUUGCGCUUCCGCC-UUUUGACAACUGCCCCUCAGUUUUCCUUUUCAUUUCUUUUUCUCGUAGAAAUGCCACCCAAAAAGCCAACCGAGAAAUCCGGUAAGGGUUUUUCAUUC .((((((..(((....))).-.))))))(((((.....)))))........(((((((.........)))))))..................(((((((((.....))))))))).... ( -27.50, z-score = -2.77, R) >droAna3.scaffold_13337 22124600 118 + 23293914 UGUCAGAUUCGGCUUCCGCU-UUUUGACAACUGCCCCUCAGUUUUCCUUUUCAUUUCUUUUUCUCGUAGAAAUGCCACCCAAAAAGCCAACCGAGAAAUCCGGUAAGGGUUUUUCAUAC .(((......)))....(((-(((((..(((((.....)))))........(((((((.........))))))).....)))))))).....(((((((((.....))))))))).... ( -27.80, z-score = -2.26, R) >dp4.chrXR_group6 7815996 118 - 13314419 UGUCACAUUACGCUUCCGCU-UUUUGACAACUGGCCCUCGGUUGUUCUUUUCAUUUCUUUUUCUCGUAGAAAUGCCACCCAAAAAGCCAACUGAGAAAUCCGGUAAGGAUUUUUCAUAU .................(((-((((((((((((.....)))))))......(((((((.........))))))).....))))))))....((((((((((.....))))))))))... ( -31.80, z-score = -3.58, R) >droPer1.super_27 562556 118 + 1201981 UGUCACAUUACGCUUCCGCU-UUUUGACAACUGGCCCUCGGUUGUUCUUUUCAUUUCUUUUUCUCGUAGAAAUGCCACCCAAAAAGCCAACUGAGAAAUCCGGUAAGGAUUUUUCAUAU .................(((-((((((((((((.....)))))))......(((((((.........))))))).....))))))))....((((((((((.....))))))))))... ( -31.80, z-score = -3.58, R) >droWil1.scaffold_180698 874604 119 - 11422946 UGUCAACCGAGACUUCCGCCUUUUUGACAACUGCCCAAUAUUUUUCCUUUUCAUUUCUUUUUCUCGUAGAAAUGCCAGCUAAAAAGCCAGCCGAAAAAUCCGGUGAGAUAAUUUAAUAG ((((((..(((........))).)))))).((..((...(((((((......((((((.........))))))((..(((....)))..)).)))))))..))..))............ ( -18.20, z-score = -0.53, R) >droVir3.scaffold_13049 2286697 119 - 25233164 UGUCAAAAUGGGCUUCCGCUCAUUUGUCAAAUGCCCCUCAGUUUUCCUUUUCAUUUCUUUUUCUCGUAGAAAUGCCACCUAAAAAGCCAACCGAGAAAUCCGGUAAGGAUUUUUCAUAU ((.((((.(((((....))))))))).))...........(((((......(((((((.........)))))))........))))).....(((((((((.....))))))))).... ( -25.94, z-score = -2.14, R) >droMoj3.scaffold_6654 986093 119 + 2564135 UGUCAAAAACUGCUUCCGCUGCUUUGUCAAAUGCCCCUUAGUUUUCCUUUUCAUUUCUUUUUCUCGUAGAAAUGCCACCUAAAAAGCCAACCGAGAAAUCCGGUAAGGAUUUUUCAUAU ((.((((....((....))...)))).))...........(((((......(((((((.........)))))))........))))).....(((((((((.....))))))))).... ( -20.24, z-score = -1.01, R) >droGri2.scaffold_15110 3323165 119 - 24565398 UGUCAGAUUUGGCUUCCGCCGCUUUGACAACUGCCCCUCAGUUUUCCUUUUCAUUUCUUUUUCUCGUAGAAAUGCCACCUAAAAAGCCAACCGAGAAGUCCGGUAAGCAUUUUUCAUAU .((((((..((((....)))).))))))(((((.....)))))........(((((((.........)))))))...........((..((((.(....)))))..))........... ( -24.30, z-score = -1.65, R) >consensus UGUCAAAUUGGGCUUCCGCC_UUUUGACAACUGCCCCUCAGUUUUCCUUUUCAUUUCUUUUUCUCGUAGAAAUGCCACCCAAAAAGCCAACCGAGAAAUCCGGUAAGGAUUUUUCAUAC (((((((....((....))...)))))))...........(((...((((((((((((.........))))))).......)))))..))).(((((((((.....))))))))).... (-19.01 = -19.55 + 0.54)

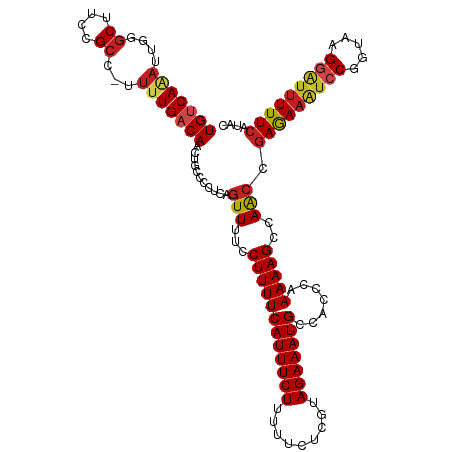
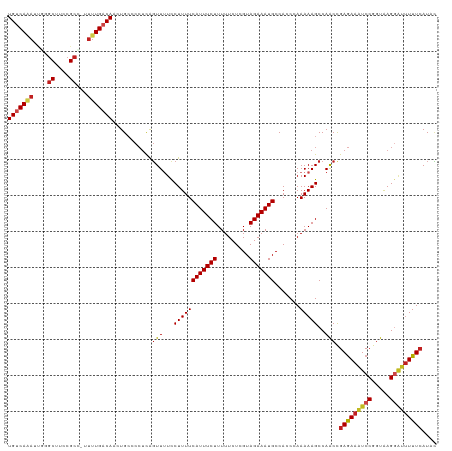
| Location | 1,668,618 – 1,668,718 |
|---|---|
| Length | 100 |
| Sequences | 12 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 81.93 |
| Shannon entropy | 0.36271 |
| G+C content | 0.46593 |
| Mean single sequence MFE | -26.96 |
| Consensus MFE | -16.62 |
| Energy contribution | -16.96 |
| Covariance contribution | 0.34 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.14 |
| Structure conservation index | 0.62 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.28 |
| SVM RNA-class probability | 0.920298 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 1668618 100 + 24543557 GGUGGCAUUUCUACGAGAAAAAGAAAUGAAAAGGAAAACUGAGGGGCAGUUGUCAAAA-GGCGGAAGCCCGUUUUGACAGGAGGGUGGCCAGAUAGCUGGA------------------ (((.((.(((((........................(((((.....)))))(((((((-(((....)))..)))))))))))).)).)))...........------------------ ( -28.30, z-score = -2.01, R) >droSim1.chr3L 1229634 100 + 22553184 GGUGGCAUUUCUACGAGAAAAAGAAAUGAAAAGGAAAACUGAGGGGCAGUUGUCAAAA-GGCGGAAGCCCGUUUUGACAGGAGGGUGGCCAGAUCGCUGGA------------------ (((.((.(((((........................(((((.....)))))(((((((-(((....)))..)))))))))))).)).)))...........------------------ ( -28.30, z-score = -1.44, R) >droSec1.super_2 1695069 100 + 7591821 GGUGGCAUUUCUACGAGAAAAAGAAAUGAAAAGGAAAACUGAGGGGCAGUUGUCAAAA-GGCGGAAGCCCGUUUUGACAGGAGGGUGGCCAGAUAGCUGGA------------------ (((.((.(((((........................(((((.....)))))(((((((-(((....)))..)))))))))))).)).)))...........------------------ ( -28.30, z-score = -2.01, R) >droYak2.chr3L 1619375 100 + 24197627 GGUGGCAUUUCUACGAGAAAAAGAAAUGAAAAGGAAAACUGAGGGGCAGUUGUCAAAA-GGCGGAAGCGCGAUUUGACAGGAGGGUGGCCAGAUCGCUGAA------------------ .....(((((((.........))))))).....((.(((((.....))))).))....-.((....))((((((((.((......))..))))))))....------------------ ( -26.10, z-score = -1.76, R) >droEre2.scaffold_4784 1648117 100 + 25762168 GGUGGCAUUUCUACGAGAAAAAGAAAUGAAAAGGAAAACUGAGGGGCAGUUGUCAAAA-GGCGGAAGCGCAAUUUGACAGGAGGGUGGCCAGAUCGCUGGA------------------ (((.((.(((((........................(((((.....)))))((((((.-.(((....)))..))))))))))).)).)))...........------------------ ( -26.50, z-score = -2.01, R) >droAna3.scaffold_13337 22124640 93 - 23293914 GGUGGCAUUUCUACGAGAAAAAGAAAUGAAAAGGAAAACUGAGGGGCAGUUGUCAAAA-AGCGGAAGCCGAAUCUGACAGUUUUUGCGCCUAAC------------------------- ((((.(((((((.........))))))).....((((((((.(((((....)))....-.((....)).....))..)))))))).))))....------------------------- ( -24.20, z-score = -1.70, R) >dp4.chrXR_group6 7816036 100 + 13314419 GGUGGCAUUUCUACGAGAAAAAGAAAUGAAAAGAACAACCGAGGGCCAGUUGUCAAAA-AGCGGAAGCGUAAUGUGACAGUAGAGUGGCCAGACCACUAAA------------------ ((((((((((((.........)))))))...............(((((.((((((...-.((....))......)))))....).)))))...)))))...------------------ ( -30.50, z-score = -3.93, R) >droPer1.super_27 562596 100 - 1201981 GGUGGCAUUUCUACGAGAAAAAGAAAUGAAAAGAACAACCGAGGGCCAGUUGUCAAAA-AGCGGAAGCGUAAUGUGACAGUAGAGUGGCCAGACCACUAAA------------------ ((((((((((((.........)))))))...............(((((.((((((...-.((....))......)))))....).)))))...)))))...------------------ ( -30.50, z-score = -3.93, R) >droWil1.scaffold_180698 874644 119 + 11422946 GCUGGCAUUUCUACGAGAAAAAGAAAUGAAAAGGAAAAAUAUUGGGCAGUUGUCAAAAAGGCGGAAGUCUCGGUUGACAGUAGAGUGGCCAGAUGUUAUCGAUAAGUGGCAACGCUAUG .(((((..((((((............((...((........))...))..((((((..((((....))))...))))))))))))..)))))............((((....))))... ( -33.50, z-score = -3.02, R) >droVir3.scaffold_13049 2286737 94 + 25233164 GGUGGCAUUUCUACGAGAAAAAGAAAUGAAAAGGAAAACUGAGGGGCAUUUGACAAAUGAGCGGAAGCCCAUUUUGACAGUAGCGUGGCCAGAU------------------------- (((.((((((((.........))))))..................((..(((.((((((.((....)).)).)))).)))..)))).)))....------------------------- ( -20.90, z-score = -0.88, R) >droMoj3.scaffold_6654 986133 94 - 2564135 GGUGGCAUUUCUACGAGAAAAAGAAAUGAAAAGGAAAACUAAGGGGCAUUUGACAAAGCAGCGGAAGCAGUUUUUGACAGUAGUGUGGCCAGAU------------------------- (((.(((((.((.((((((....(((((...((.....))......))))).........((....))..))))))..)).))))).)))....------------------------- ( -19.80, z-score = -1.03, R) >droGri2.scaffold_15110 3323205 94 + 24565398 GGUGGCAUUUCUACGAGAAAAAGAAAUGAAAAGGAAAACUGAGGGGCAGUUGUCAAAGCGGCGGAAGCCAAAUCUGACAGUGGCGUGGCCAGAU------------------------- (((.((((((((.........))))))..................((..((((((.(..(((....)))...).))))))..)))).)))....------------------------- ( -26.60, z-score = -2.00, R) >consensus GGUGGCAUUUCUACGAGAAAAAGAAAUGAAAAGGAAAACUGAGGGGCAGUUGUCAAAA_GGCGGAAGCCCAAUUUGACAGUAGGGUGGCCAGAUCGCUG_A__________________ (((.((((((((.........))))))......................((((((.....((....))......))))))....)).)))............................. (-16.62 = -16.96 + 0.34)
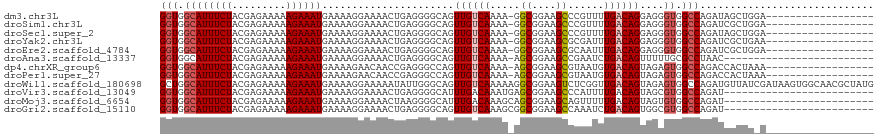
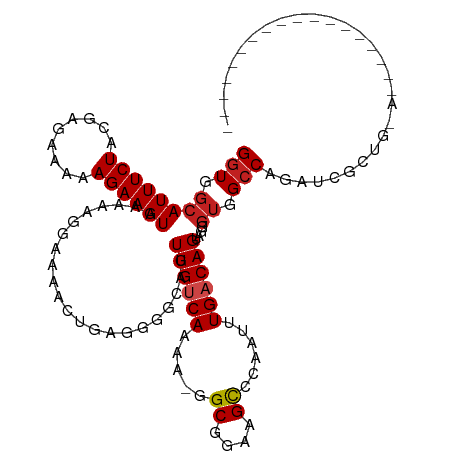
| Location | 1,668,618 – 1,668,718 |
|---|---|
| Length | 100 |
| Sequences | 12 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 81.93 |
| Shannon entropy | 0.36271 |
| G+C content | 0.46593 |
| Mean single sequence MFE | -18.77 |
| Consensus MFE | -12.45 |
| Energy contribution | -12.93 |
| Covariance contribution | 0.47 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.89 |
| Structure conservation index | 0.66 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.07 |
| SVM RNA-class probability | 0.885292 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
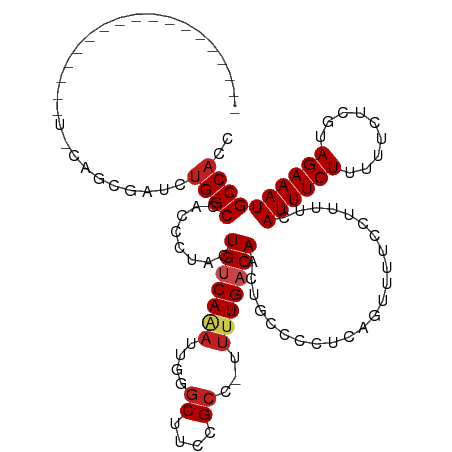
>dm3.chr3L 1668618 100 - 24543557 ------------------UCCAGCUAUCUGGCCACCCUCCUGUCAAAACGGGCUUCCGCC-UUUUGACAACUGCCCCUCAGUUUUCCUUUUCAUUUCUUUUUCUCGUAGAAAUGCCACC ------------------.((((....))))..........((((((..((((....)))-)))))))(((((.....)))))........(((((((.........)))))))..... ( -20.40, z-score = -2.50, R) >droSim1.chr3L 1229634 100 - 22553184 ------------------UCCAGCGAUCUGGCCACCCUCCUGUCAAAACGGGCUUCCGCC-UUUUGACAACUGCCCCUCAGUUUUCCUUUUCAUUUCUUUUUCUCGUAGAAAUGCCACC ------------------.((((....))))..........((((((..((((....)))-)))))))(((((.....)))))........(((((((.........)))))))..... ( -20.40, z-score = -1.91, R) >droSec1.super_2 1695069 100 - 7591821 ------------------UCCAGCUAUCUGGCCACCCUCCUGUCAAAACGGGCUUCCGCC-UUUUGACAACUGCCCCUCAGUUUUCCUUUUCAUUUCUUUUUCUCGUAGAAAUGCCACC ------------------.((((....))))..........((((((..((((....)))-)))))))(((((.....)))))........(((((((.........)))))))..... ( -20.40, z-score = -2.50, R) >droYak2.chr3L 1619375 100 - 24197627 ------------------UUCAGCGAUCUGGCCACCCUCCUGUCAAAUCGCGCUUCCGCC-UUUUGACAACUGCCCCUCAGUUUUCCUUUUCAUUUCUUUUUCUCGUAGAAAUGCCACC ------------------..........((((.........((((((..(((....))).-.))))))(((((.....))))).........((((((.........)))))))))).. ( -19.10, z-score = -2.71, R) >droEre2.scaffold_4784 1648117 100 - 25762168 ------------------UCCAGCGAUCUGGCCACCCUCCUGUCAAAUUGCGCUUCCGCC-UUUUGACAACUGCCCCUCAGUUUUCCUUUUCAUUUCUUUUUCUCGUAGAAAUGCCACC ------------------.((((....))))..........((((((..(((....))).-.))))))(((((.....)))))........(((((((.........)))))))..... ( -19.50, z-score = -2.89, R) >droAna3.scaffold_13337 22124640 93 + 23293914 -------------------------GUUAGGCGCAAAAACUGUCAGAUUCGGCUUCCGCU-UUUUGACAACUGCCCCUCAGUUUUCCUUUUCAUUUCUUUUUCUCGUAGAAAUGCCACC -------------------------...(((.(((.....(((((((...(((....)))-.)))))))..))).))).............(((((((.........)))))))..... ( -18.10, z-score = -1.45, R) >dp4.chrXR_group6 7816036 100 - 13314419 ------------------UUUAGUGGUCUGGCCACUCUACUGUCACAUUACGCUUCCGCU-UUUUGACAACUGGCCCUCGGUUGUUCUUUUCAUUUCUUUUUCUCGUAGAAAUGCCACC ------------------....((((...(((((......(((((......((....)).-...)))))..)))))...............(((((((.........))))))))))). ( -20.20, z-score = -1.52, R) >droPer1.super_27 562596 100 + 1201981 ------------------UUUAGUGGUCUGGCCACUCUACUGUCACAUUACGCUUCCGCU-UUUUGACAACUGGCCCUCGGUUGUUCUUUUCAUUUCUUUUUCUCGUAGAAAUGCCACC ------------------....((((...(((((......(((((......((....)).-...)))))..)))))...............(((((((.........))))))))))). ( -20.20, z-score = -1.52, R) >droWil1.scaffold_180698 874644 119 - 11422946 CAUAGCGUUGCCACUUAUCGAUAACAUCUGGCCACUCUACUGUCAACCGAGACUUCCGCCUUUUUGACAACUGCCCAAUAUUUUUCCUUUUCAUUUCUUUUUCUCGUAGAAAUGCCAGC ..(((.((.((((...............)))).)).))).((((((..(((........))).))))))......................(((((((.........)))))))..... ( -16.96, z-score = -0.69, R) >droVir3.scaffold_13049 2286737 94 - 25233164 -------------------------AUCUGGCCACGCUACUGUCAAAAUGGGCUUCCGCUCAUUUGUCAAAUGCCCCUCAGUUUUCCUUUUCAUUUCUUUUUCUCGUAGAAAUGCCACC -------------------------...((((...((...((.((((.(((((....))))))))).))...))..................((((((.........)))))))))).. ( -18.80, z-score = -2.21, R) >droMoj3.scaffold_6654 986133 94 + 2564135 -------------------------AUCUGGCCACACUACUGUCAAAAACUGCUUCCGCUGCUUUGUCAAAUGCCCCUUAGUUUUCCUUUUCAUUUCUUUUUCUCGUAGAAAUGCCACC -------------------------...((((........((.((((....((....))...)))).)).......................((((((.........)))))))))).. ( -11.00, z-score = -0.39, R) >droGri2.scaffold_15110 3323205 94 - 24565398 -------------------------AUCUGGCCACGCCACUGUCAGAUUUGGCUUCCGCCGCUUUGACAACUGCCCCUCAGUUUUCCUUUUCAUUUCUUUUUCUCGUAGAAAUGCCACC -------------------------...((((...))))..((((((..((((....)))).))))))(((((.....)))))........(((((((.........)))))))..... ( -20.20, z-score = -2.34, R) >consensus __________________U_CAGCGAUCUGGCCACCCUACUGUCAAAUUGGGCUUCCGCC_UUUUGACAACUGCCCCUCAGUUUUCCUUUUCAUUUCUUUUUCUCGUAGAAAUGCCACC ............................((((........(((((((....((....))...))))))).......................((((((.........)))))))))).. (-12.45 = -12.93 + 0.47)
| Location | 1,668,658 – 1,668,752 |
|---|---|
| Length | 94 |
| Sequences | 12 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 61.29 |
| Shannon entropy | 0.74300 |
| G+C content | 0.53831 |
| Mean single sequence MFE | -29.73 |
| Consensus MFE | -10.81 |
| Energy contribution | -11.07 |
| Covariance contribution | 0.26 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.54 |
| Structure conservation index | 0.36 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.23 |
| SVM RNA-class probability | 0.913468 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 1668658 94 + 24543557 GAGGGGCAGUUGUCAAAA-GGCGGAAGCCCGUUUUGACAGGAGGGUGGCCAGAUA--GCUGGAAUUU-GGCACGCAUUCAGUGCGUUUGGCAACACUG---------------- .(((.(((.(((((((((-(((....)))..)))))))))(((.((((((((((.--......))))-))).))).)))..))).)))(....)....---------------- ( -34.10, z-score = -1.95, R) >droSim1.chr3L 1229674 94 + 22553184 GAGGGGCAGUUGUCAAAA-GGCGGAAGCCCGUUUUGACAGGAGGGUGGCCAGAUC--GCUGGAAUUU-UGCACGCAUUCAGUGCGCUUGGCAACACUG---------------- .........(((((((((-(((....)))..)))))))))...((((.((((...--.))))....(-(((.(((((...)))))....)))))))).---------------- ( -31.80, z-score = -0.59, R) >droSec1.super_2 1695109 94 + 7591821 GAGGGGCAGUUGUCAAAA-GGCGGAAGCCCGUUUUGACAGGAGGGUGGCCAGAUA--GCUGGAAUUU-UGCACGCAAUCAGUGCGCUUGGCAACACUG---------------- (..((((..(((((((((-(((....)))..))))))))).(((((..((((...--.)))).))))-)((((.......))))))))..).......---------------- ( -31.50, z-score = -0.81, R) >droYak2.chr3L 1619415 88 + 24197627 GAGGGGCAGUUGUCAAAA-GGCGGAAGCGCGAUUUGACAGGAGGGUGGCCAGAUC--GCUGAAAUUUCGGCACGCAUUC-GUGCGCUUGGCA---------------------- .........((((((((.-.(((....)))..)))))))).......(((((...--(((((....))))).((((...-.)))).))))).---------------------- ( -29.70, z-score = -0.66, R) >droEre2.scaffold_4784 1648157 94 + 25762168 GAGGGGCAGUUGUCAAAA-GGCGGAAGCGCAAUUUGACAGGAGGGUGGCCAGAUC--GCUGGAAUUU-GGCACGCAUUCAGUGCGCUUGGCAAUACUC---------------- .(((.(((.((((((((.-.(((....)))..))))))))(((.(((((((((((--....).))))-))).))).)))..))).)))..........---------------- ( -33.60, z-score = -2.27, R) >droAna3.scaffold_13337 22124680 80 - 23293914 GAGGGGCAGUUGUCAAAA-AGCGGAAGCCGAAUCUGACAGUUUUUGCGCCUAACA--GCACGCGUCU-------------GCGCGCUUAACAGUGU------------------ .(((.((((((((((...-.((....))......))))))...)))).)))...(--((.(((....-------------))).))).........------------------ ( -24.90, z-score = -0.07, R) >dp4.chrXR_group6 7816076 81 + 13314419 GAGGGCCAGUUGUCAAAA-AGCGGAAGCGUAAUGUGACAGUAGAGUGGCCAGACC--ACUAAAUAUU--------------UGCACUUGACAGCACUG---------------- ........((((((((..-.((....)).....(((..((((.(((((.....))--)))...))))--------------..)))))))))))....---------------- ( -25.80, z-score = -2.47, R) >droPer1.super_27 562636 81 - 1201981 GAGGGCCAGUUGUCAAAA-AGCGGAAGCGUAAUGUGACAGUAGAGUGGCCAGACC--ACUAAAUAUU--------------UGCACUUGACAGCACUG---------------- ........((((((((..-.((....)).....(((..((((.(((((.....))--)))...))))--------------..)))))))))))....---------------- ( -25.80, z-score = -2.47, R) >droWil1.scaffold_180698 874684 114 + 11422946 AUUGGGCAGUUGUCAAAAAGGCGGAAGUCUCGGUUGACAGUAGAGUGGCCAGAUGUUAUCGAUAAGUGGCAACGCUAUGCGUUAACAGCACACACUGUGUGCGCUUAACAGCAC .((((.((.(((((((..((((....))))...))))))....).)).)))).(((((.((...((((....))))...)).)))))((((((...))))))(((....))).. ( -41.80, z-score = -2.86, R) >droVir3.scaffold_13049 2286777 78 + 25233164 GAGGGGCAUUUGACAAAUGAGCGGAAGCCCAUUUUGACAGUAGCGUGGCCAGAUG-CGU-AGCAAGUGGCAACGCUGUAU---------------------------------- ...((((.((((.(......))))).))))..........((((((.((((..((-(..-.)))..)))).))))))...---------------------------------- ( -26.30, z-score = -1.84, R) >droMoj3.scaffold_6654 986173 78 - 2564135 AAGGGGCAUUUGACAAAGCAGCGGAAGCAGUUUUUGACAGUAGUGUGGCCAGAUG-GGU-GGCAAGUGGCAACGCUCAAA---------------------------------- ...((((...((.((((((.((....)).).))))).))((..(((.(((.....-)))-.)))....))...))))...---------------------------------- ( -21.80, z-score = -0.67, R) >droGri2.scaffold_15110 3323245 75 + 24565398 GAGGGGCAGUUGUCAAAGCGGCGGAAGCCAAAUCUGACAGUGGCGUGGCCAGAUGGUGU--GCGGGUGGCAACGCUC------------------------------------- ...((((.(((((((..(((((....)))........((.((((...))))..))....--))...)))))))))))------------------------------------- ( -29.60, z-score = -1.85, R) >consensus GAGGGGCAGUUGUCAAAA_GGCGGAAGCCCAAUUUGACAGUAGGGUGGCCAGAUC__GCUGGAAUUU_GGCACGCAUUC_GUGCGCUUGGCA_CACU_________________ ...((.((.((((((.....((....))......)))))).....)).))................................................................ (-10.81 = -11.07 + 0.26)

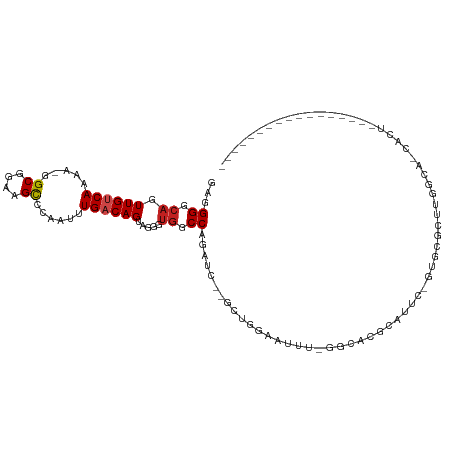
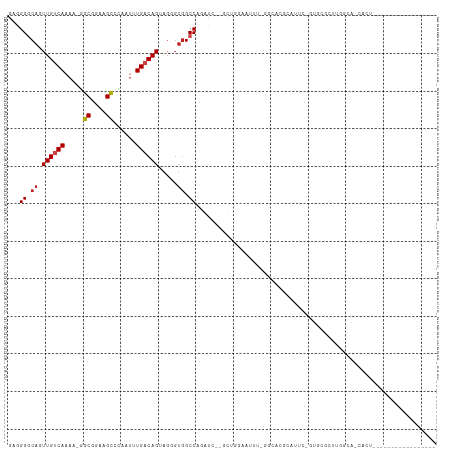
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:56:10 2011