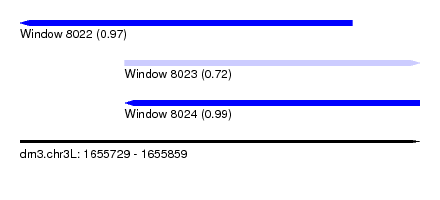
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 1,655,729 – 1,655,859 |
| Length | 130 |
| Max. P | 0.991533 |

| Location | 1,655,729 – 1,655,837 |
|---|---|
| Length | 108 |
| Sequences | 12 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 84.08 |
| Shannon entropy | 0.32781 |
| G+C content | 0.48202 |
| Mean single sequence MFE | -29.20 |
| Consensus MFE | -23.92 |
| Energy contribution | -23.41 |
| Covariance contribution | -0.51 |
| Combinations/Pair | 1.22 |
| Mean z-score | -1.96 |
| Structure conservation index | 0.82 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.78 |
| SVM RNA-class probability | 0.967125 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
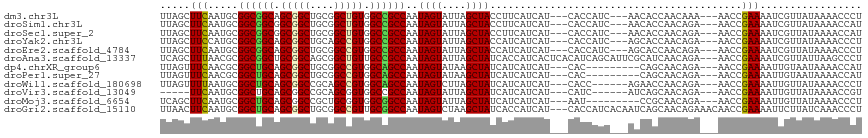
>dm3.chr3L 1655729 108 - 24543557 UUAGCUUCAAUGCGGCGGCAGCGGCUGCGGCUGUGGCCGCCAAUAGUAUUAGCUACCUUCAUCAU---CACCAUC---AACACCAACAAA---AACCGAAAAUCGUUAUAAAACCCU .(((((..(((((((((((.(((((....))))).))))))....))))))))))..........---.......---............---........................ ( -27.50, z-score = -1.51, R) >droSim1.chr3L 1216381 108 - 22553184 UUAGCUUCAAUGCGGCGGCGGCGGCUGCGGCUGUGGCCGCCAAUAGUAUUAGCUACCUUCAUCAU---CACCAUC---AACACCAACAGA---AACCGAAAAUCGUUAUAAAACCAU .(((((..(((((((((((.(((((....))))).))))))....))))))))))..........---.......---............---........................ ( -29.60, z-score = -1.43, R) >droSec1.super_2 1681919 108 - 7591821 UUAGCUUCAAUGCGGCGGCGGCGGCUGCGGCUGUGGCCGCCAAUAGUAUUAGCUACCUUCAUCAU---CACCAUC---AACACCAACAGA---AACCGAAAAUCGUUAUAAAACCAU .(((((..(((((((((((.(((((....))))).))))))....))))))))))..........---.......---............---........................ ( -29.60, z-score = -1.43, R) >droYak2.chr3L 1606397 108 - 24197627 UUAGCUUCCAUGCGGCGGCAGCGGCUGCAGCCGUGGCCGCCAAUAGUAUUAGCUACCAUCAUCAU---CACCAUC---AGCACCAACAGA---AACCGAAAAUCGUUAUAAAACCCU .(((((...((((((((((.(((((....))))).))))))....)))).)))))..........---.......---............---........................ ( -29.10, z-score = -2.01, R) >droEre2.scaffold_4784 1635315 108 - 25762168 UUAGCUUCAAUGCGGCGGCAGCGGCUGCGGCCGUGGCCGCCAAUAGUAUUAGCUACCAUCAUCAU---CACCAUC---AGCACCAACAGA---AACCGAAAAUCGUUAUAAAACCCU .(((((..(((((((((((.(((((....))))).))))))....))))))))))..........---.......---............---........................ ( -29.70, z-score = -1.56, R) >droAna3.scaffold_13337 22111888 114 + 23293914 UCAGCUUUAACGCGGCGGCUGCGGCAGCGGCUGUUGCCGCCAAUAGUAUUAGCUAUCACCAUCACUCACAUCAGCAUUCGCAUCAACAGA---AACCGAAAAUCGUUAUUAAGCCCU ...((((((((((((((((.(((((....))))).)))))).(((((....))))).................)).((((..((....))---...))))....))))..))))... ( -33.40, z-score = -2.24, R) >dp4.chrXR_group6 7802014 102 - 13314419 UUAGUUUCAACGCGGCUGCAGCGGCUGCGGCCGUGGCAGCCAAUAGUAUAAGCUAUCAUCAUCAU---CAC---------CAGCAACAGA---AACCGAAAAUUGUAAUAAAACCAU (..(((((...((((((((.(((((....))))).)))))).(((((....))))).........---...---------..))....))---)))..).................. ( -30.40, z-score = -2.84, R) >droPer1.super_27 548429 102 + 1201981 UUAGUUUCAACGCGGCUGCAGCGGCUGCGGCCGUGGCAGCCAAUAGUAUAAGCUAUCAUCAUCAU---CAC---------CAGCAACAGA---AACCGAAAAUUGUAAUAAAACCAU (..(((((...((((((((.(((((....))))).)))))).(((((....))))).........---...---------..))....))---)))..).................. ( -30.40, z-score = -2.84, R) >droWil1.scaffold_180698 859279 105 - 11422946 UUAGUUUUAAUGCGGCUGCAGCGGCCGCAGCCGUGGCAGCCAAUAGUCUUAGCUAUCAUCAUCAU---CACC------AGAACCAACAGA---AACCGAAAAUUGUUAUAAAACCCU ...((((((..((((((((.(((((....))))).)))))).(((((....))))).........---....------............---...........))..))))))... ( -27.60, z-score = -2.44, R) >droVir3.scaffold_13049 2272537 100 - 25233164 -----UUCAAUGCGGCUGCAGCGGCCGCAGCGGUGGCCGCCAAUAGUAUUAGCUAUCAUCAUCAU---CAUC------AUCAGCAACAGA---AACCGAAAAUUGUUAUAAAACCGU -----.....((..((((..((((((((....))))))))..(((((....))))).........---....------..))))..))..---........................ ( -25.20, z-score = -1.04, R) >droMoj3.scaffold_6654 971945 102 + 2564135 UCAGCUUCAAUGCGGCUGCAGCGGCCGCUGCGGUGGCGGCCAAUAGUAUUAGCUAUCAUCAUCAU---AAU---------CCGCAACAGA---AACCGAAAAUUGUUAUAAAACCCU .......((((.((((((..(((((((((.....))))))).(((((....))))).........---...---------..))..))).---..)))...))))............ ( -26.80, z-score = -0.45, R) >droGri2.scaffold_15110 3309052 114 - 24565398 UUAACUUCAAUGCGGCUGCAGCGGCUGCGGCCGUUGCGGCCAAUAGUCUAAGCUAUCACCAUCAU---CACCAUCACAAUCAGCAACAGAAACAACCGAAAAUUCUUAUCAAACCCU .....(((..(((((((((((((((....)))))))))))).(((((....))))).........---..............)))...))).......................... ( -31.10, z-score = -3.70, R) >consensus UUAGCUUCAAUGCGGCGGCAGCGGCUGCGGCCGUGGCCGCCAAUAGUAUUAGCUAUCAUCAUCAU___CACCAUC___AGCACCAACAGA___AACCGAAAAUCGUUAUAAAACCCU .....(((.....((((((.(((((....))))).))))))..((((....))))..........................................)))................. (-23.92 = -23.41 + -0.51)
| Location | 1,655,763 – 1,655,859 |
|---|---|
| Length | 96 |
| Sequences | 13 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 68.14 |
| Shannon entropy | 0.61017 |
| G+C content | 0.58408 |
| Mean single sequence MFE | -31.89 |
| Consensus MFE | -16.16 |
| Energy contribution | -15.65 |
| Covariance contribution | -0.51 |
| Combinations/Pair | 1.74 |
| Mean z-score | -0.92 |
| Structure conservation index | 0.51 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.49 |
| SVM RNA-class probability | 0.716982 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
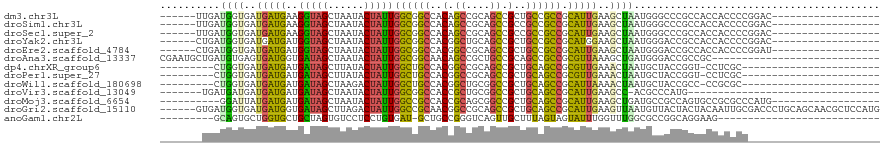
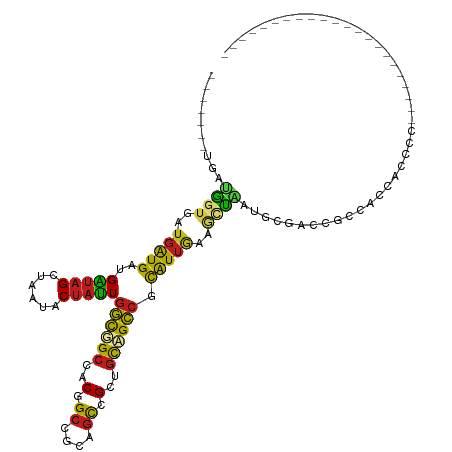
>dm3.chr3L 1655763 96 + 24543557 ------UUGAUGGUGAUGAUGAAGGUAGCUAAUACUAUUGGCGGCCACAGCCGCAGCCGCUGCCGCCGCAUUGAAGCUAAUGGGCCCGCCACCACCCCGGAC------------------ ------.((.(((((........((((((.....(....)(((((....)))))....))))))(((.(((((....)))))))).))))).))........------------------ ( -33.50, z-score = -0.71, R) >droSim1.chr3L 1216415 96 + 22553184 ------UUGAUGGUGAUGAUGAAGGUAGCUAAUACUAUUGGCGGCCACAGCCGCAGCCGCCGCCGCCGCAUUGAAGCUAAUGGGCCCGCCACCACCCCGGAC------------------ ------....(((((........((..(((....(....)(((((....))))))))..))((.(((.(((((....))))))))..)))))))........------------------ ( -31.30, z-score = 0.01, R) >droSec1.super_2 1681953 96 + 7591821 ------UUGAUGGUGAUGAUGAAGGUAGCUAAUACUAUUGGCGGCCACAGCCGCAGCCGCCGCCGCCGCAUUGAAGCUAAUGGGCCCGCCACCACCCCGGAC------------------ ------....(((((........((..(((....(....)(((((....))))))))..))((.(((.(((((....))))))))..)))))))........------------------ ( -31.30, z-score = 0.01, R) >droYak2.chr3L 1606431 96 + 24197627 ------CUGAUGGUGAUGAUGAUGGUAGCUAAUACUAUUGGCGGCCACGGCUGCAGCCGCUGCCGCCGCAUGGAAGCUAAUGGGACCGCCACCACCCCGGAC------------------ ------(((..((((.....(((((((.....)))))))(((((((.((((.((((...)))).))))(((........))))).)))))..)))).)))..------------------ ( -37.80, z-score = -1.72, R) >droEre2.scaffold_4784 1635349 96 + 25762168 ------CUGAUGGUGAUGAUGAUGGUAGCUAAUACUAUUGGCGGCCACGGCCGCAGCCGCUGCCGCCGCAUUGAAGCUAAUGGGACCGCCACCACCCCGGAU------------------ ------(((..((((.....(((((((.....)))))))(((((((.((((.((((...)))).))))(((((....))))))).)))))..)))).)))..------------------ ( -38.80, z-score = -2.12, R) >droAna3.scaffold_13337 22111922 92 - 23293914 CGAAUGCUGAUGUGAGUGAUGGUGAUAGCUAAUACUAUUGGCGGCAACAGCCGCUGCCGCAGCCGCCGCGUUAAAGCUGAUGGGACCGCCGC---------------------------- .....(((((((((.(((.(((((((((......)))))((((((....))))))))))....)))))))))..)))....((.....))..---------------------------- ( -26.90, z-score = 1.14, R) >dp4.chrXR_group6 7802045 87 + 13314419 ---------CUGGUGAUGAUGAUGAUAGCUUAUACUAUUGGCUGCCACGGCCGCAGCCGCUGCAGCCGCGUUGAAACUAAUGCUACCGGU-CCUCGC----------------------- ---------((((((........(((((......)))))((((((..((((....))))..))))))((((((....)))))))))))).-......----------------------- ( -31.70, z-score = -2.08, R) >droPer1.super_27 548460 87 - 1201981 ---------CUGGUGAUGAUGAUGAUAGCUUAUACUAUUGGCUGCCACGGCCGCAGCCGCUGCAGCCGCGUUGAAACUAAUGCUACCGGU-CCUCGC----------------------- ---------((((((........(((((......)))))((((((..((((....))))..))))))((((((....)))))))))))).-......----------------------- ( -31.70, z-score = -2.08, R) >droWil1.scaffold_180698 859313 87 + 11422946 ---------CUGGUGAUGAUGAUGAUAGCUAAGACUAUUGGCUGCCACGGCUGCGGCCGCUGCAGCCGCAUUAAAACUAAUGCUACCGCC-CCGCGC----------------------- ---------..((((........(((((......)))))((((((..((((....))))..))))))((((((....))))))...))))-......----------------------- ( -31.60, z-score = -2.16, R) >droVir3.scaffold_13049 2272569 80 + 25233164 -------UGAUGAUGAUGAUGAUGAUAGCUAAUACUAUUGGCGGCCACCGCUGCGGCCGCUGCAGCCGCAUUGAAGCC-ACGCCCAUG-------------------------------- -------...........(((..(((((......)))))((((((...((.((((((.......)))))).))..)))-..)))))).-------------------------------- ( -21.00, z-score = 0.34, R) >droMoj3.scaffold_6654 971977 92 - 2564135 ----------GGAUUAUGAUGAUGAUAGCUAAUACUAUUGGCCGCCACCGCAGCGGCCGCUGCAGCCGCAUUGAAGCUGAUGCCGCCAGUGCCGCGCCCAUG------------------ ----------....((((.....(((((......)))))(((.((....)).(((((.((((..((.(((((......))))).))))))))))))))))))------------------ ( -29.40, z-score = 0.07, R) >droGri2.scaffold_15110 3309092 114 + 24565398 ------GUGAUGGUGAUGAUGGUGAUAGCUUAGACUAUUGGCCGCAACGGCCGCAGCCGCUGCAGCCGCAUUGAAGUUAAUGUUACUACUACAAUUGCGACCCUGCAGCAACGCUCCAUG ------...((((.(.((..((((((((......)))))(((((...)))))...)))(((((((.(((((((.(((..........))).))).))))...)))))))..))).)))). ( -38.40, z-score = -1.56, R) >anoGam1.chr2L 23821101 83 - 48795086 ---------GCAGUGCUGGUGCUGCUAGUGUCCUCCUGUGAU-GCUGCCGGGUCAGUUGCUUUAGUAGUAUUUGGUUUGGCGCCGGCAGGAAG--------------------------- ---------((((((....))))))........((((((...-((.(((((..(((.((((.....)))).)))..)))))))..))))))..--------------------------- ( -31.20, z-score = -1.05, R) >consensus _______UGAUGGUGAUGAUGAUGAUAGCUAAUACUAUUGGCGGCCACGGCCGCAGCCGCUGCAGCCGCAUUGAAGCUAAUGCGACCGCCACCACCCC______________________ ..........((((..(((((..(((((......)))))((((((..(.((....)).)..)))))).)))))..))))......................................... (-16.16 = -15.65 + -0.51)
| Location | 1,655,763 – 1,655,859 |
|---|---|
| Length | 96 |
| Sequences | 13 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 68.14 |
| Shannon entropy | 0.61017 |
| G+C content | 0.58408 |
| Mean single sequence MFE | -35.80 |
| Consensus MFE | -20.40 |
| Energy contribution | -20.48 |
| Covariance contribution | 0.08 |
| Combinations/Pair | 1.47 |
| Mean z-score | -1.73 |
| Structure conservation index | 0.57 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.48 |
| SVM RNA-class probability | 0.991533 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
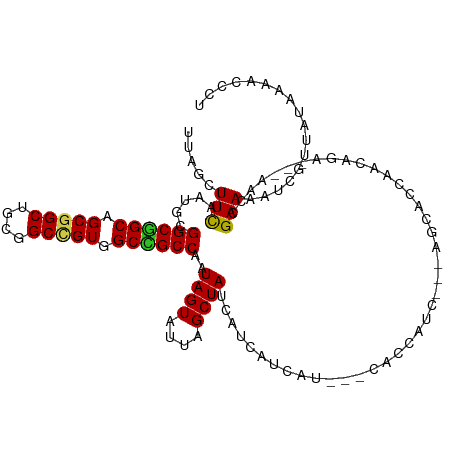
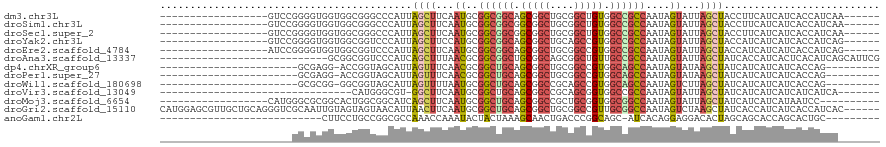
>dm3.chr3L 1655763 96 - 24543557 ------------------GUCCGGGGUGGUGGCGGGCCCAUUAGCUUCAAUGCGGCGGCAGCGGCUGCGGCUGUGGCCGCCAAUAGUAUUAGCUACCUUCAUCAUCACCAUCAA------ ------------------....((((((((((.(((.....(((((..(((((((((((.(((((....))))).))))))....))))))))))))))))))))).)).....------ ( -41.20, z-score = -1.53, R) >droSim1.chr3L 1216415 96 - 22553184 ------------------GUCCGGGGUGGUGGCGGGCCCAUUAGCUUCAAUGCGGCGGCGGCGGCUGCGGCUGUGGCCGCCAAUAGUAUUAGCUACCUUCAUCAUCACCAUCAA------ ------------------....((((((((((.(((.....(((((..(((((((((((.(((((....))))).))))))....))))))))))))))))))))).)).....------ ( -43.30, z-score = -1.82, R) >droSec1.super_2 1681953 96 - 7591821 ------------------GUCCGGGGUGGUGGCGGGCCCAUUAGCUUCAAUGCGGCGGCGGCGGCUGCGGCUGUGGCCGCCAAUAGUAUUAGCUACCUUCAUCAUCACCAUCAA------ ------------------....((((((((((.(((.....(((((..(((((((((((.(((((....))))).))))))....))))))))))))))))))))).)).....------ ( -43.30, z-score = -1.82, R) >droYak2.chr3L 1606431 96 - 24197627 ------------------GUCCGGGGUGGUGGCGGUCCCAUUAGCUUCCAUGCGGCGGCAGCGGCUGCAGCCGUGGCCGCCAAUAGUAUUAGCUACCAUCAUCAUCACCAUCAG------ ------------------....(((((((((..(((......((((...((((((((((.(((((....))))).))))))....)))).)))))))..))))))).)).....------ ( -43.10, z-score = -3.04, R) >droEre2.scaffold_4784 1635349 96 - 25762168 ------------------AUCCGGGGUGGUGGCGGUCCCAUUAGCUUCAAUGCGGCGGCAGCGGCUGCGGCCGUGGCCGCCAAUAGUAUUAGCUACCAUCAUCAUCACCAUCAG------ ------------------....(((((((((..(((......((((..(((((((((((.(((((....))))).))))))....))))))))))))..))))))).)).....------ ( -43.70, z-score = -3.04, R) >droAna3.scaffold_13337 22111922 92 + 23293914 ----------------------------GCGGCGGUCCCAUCAGCUUUAACGCGGCGGCUGCGGCAGCGGCUGUUGCCGCCAAUAGUAUUAGCUAUCACCAUCACUCACAUCAGCAUUCG ----------------------------((((((((.......((......))(((((((((....))))))))))))))).(((((....))))).................))..... ( -31.50, z-score = -1.54, R) >dp4.chrXR_group6 7802045 87 - 13314419 -----------------------GCGAGG-ACCGGUAGCAUUAGUUUCAACGCGGCUGCAGCGGCUGCGGCCGUGGCAGCCAAUAGUAUAAGCUAUCAUCAUCAUCACCAG--------- -----------------------..((.(-(..((((((....((......))((((((.(((((....))))).))))))..........)))))).)).))........--------- ( -32.30, z-score = -2.32, R) >droPer1.super_27 548460 87 + 1201981 -----------------------GCGAGG-ACCGGUAGCAUUAGUUUCAACGCGGCUGCAGCGGCUGCGGCCGUGGCAGCCAAUAGUAUAAGCUAUCAUCAUCAUCACCAG--------- -----------------------..((.(-(..((((((....((......))((((((.(((((....))))).))))))..........)))))).)).))........--------- ( -32.30, z-score = -2.32, R) >droWil1.scaffold_180698 859313 87 - 11422946 -----------------------GCGCGG-GGCGGUAGCAUUAGUUUUAAUGCGGCUGCAGCGGCCGCAGCCGUGGCAGCCAAUAGUCUUAGCUAUCAUCAUCAUCACCAG--------- -----------------------((..((-(((....((((((....))))))((((((.(((((....))))).))))))....))))).))..................--------- ( -34.20, z-score = -2.28, R) >droVir3.scaffold_13049 2272569 80 - 25233164 --------------------------------CAUGGGCGU-GGCUUCAAUGCGGCUGCAGCGGCCGCAGCGGUGGCCGCCAAUAGUAUUAGCUAUCAUCAUCAUCAUCAUCA------- --------------------------------.((((..((-((((..(((((...((..((((((((....))))))))))...)))))))))))..))))...........------- ( -27.00, z-score = -0.57, R) >droMoj3.scaffold_6654 971977 92 + 2564135 ------------------CAUGGGCGCGGCACUGGCGGCAUCAGCUUCAAUGCGGCUGCAGCGGCCGCUGCGGUGGCGGCCAAUAGUAUUAGCUAUCAUCAUCAUAAUCC---------- ------------------.((((((.((.(((((.((((....((......))(((((...)))))))))))))).))))).(((((....)))))......))).....---------- ( -34.80, z-score = -0.45, R) >droGri2.scaffold_15110 3309092 114 - 24565398 CAUGGAGCGUUGCUGCAGGGUCGCAAUUGUAGUAGUAACAUUAACUUCAAUGCGGCUGCAGCGGCUGCGGCCGUUGCGGCCAAUAGUCUAAGCUAUCACCAUCAUCACCAUCAC------ .((((.((((((((((((........)))))).(((.......))).))))))((((((((((((....)))))))))))).(((((....)))))..))))............------ ( -40.70, z-score = -1.94, R) >anoGam1.chr2L 23821101 83 + 48795086 ---------------------------CUUCCUGCCGGCGCCAAACCAAAUACUACUAAAGCAACUGACCCGGCAGC-AUCACAGGAGGACACUAGCAGCACCAGCACUGC--------- ---------------------------(((((((...(((((.............................))).))-....)))))))......((((........))))--------- ( -17.95, z-score = 0.13, R) >consensus ______________________GGGGUGGUGGCGGUCCCAUUAGCUUCAAUGCGGCUGCAGCGGCUGCGGCCGUGGCCGCCAAUAGUAUUAGCUAUCAUCAUCAUCACCAUCA_______ ..........................................((((...((..((((((.(((((....))))).))))))....))...)))).......................... (-20.40 = -20.48 + 0.08)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:56:04 2011