| Sequence ID | dm3.chr3L |
|---|---|
| Location | 1,632,874 – 1,632,965 |
| Length | 91 |
| Max. P | 0.999173 |

| Location | 1,632,874 – 1,632,965 |
|---|---|
| Length | 91 |
| Sequences | 6 |
| Columns | 96 |
| Reading direction | reverse |
| Mean pairwise identity | 64.24 |
| Shannon entropy | 0.68139 |
| G+C content | 0.46573 |
| Mean single sequence MFE | -23.17 |
| Consensus MFE | -9.13 |
| Energy contribution | -10.35 |
| Covariance contribution | 1.23 |
| Combinations/Pair | 1.46 |
| Mean z-score | -2.99 |
| Structure conservation index | 0.39 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.69 |
| SVM RNA-class probability | 0.999173 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 1632874 91 - 24543557 -UGCUCCCACU-UCCGUUUCCGCCGUUCCUCUGGUUUUAUAUUAUUUC-GGCUGGCUGUAAACGGUGUCUCAUC--GUUUACCGCCAGCCUUAGCC -.(((......-.........((((......)))).............-(((((((.((((((((((...))))--)))))).)))))))..))). ( -31.40, z-score = -5.73, R) >dp4.chrXR_group8 4190494 91 - 9212921 CCGUUUCCGCUGUCCUGUUACGCUG-UCUUCUGUUACCGU-UAGCGUUAGCGUUACCGU-UGCCCCCCGUUAAC--GUUUACUUAUAAUUUUAGCU ........((((....((((.((((-..............-))))((.(((((((.((.-.......)).))))--))).))...))))..)))). ( -15.54, z-score = -1.09, R) >droEre2.scaffold_4784 1612448 92 - 25762168 -UGCUACCACU-UCCUUUUCCGCUGUUCCCCUGAUUUUAUAUGAUUUCAGGCUGACUGUAAGCGGUGCCUCAUC--GUUUACGGCCAGCUUUAGCC -..........-.........((((.....((((..((....))..))))((((.((((((((((((...))))--)))))))).))))..)))). ( -28.20, z-score = -3.74, R) >droYak2.chr3L 1584061 92 - 24197627 -UGCUCCCACU-UCCGUUUCCGCUGUUCCCCUGGUUUUAUAUGAUUUCAGGCUGGCUGUAAGCGGUUCCUCAUC--GUUUACCGCCAGCUUUAGCC -..........-.........((((.....((((..((....))..))))((((((.(((((((((.....)))--)))))).))))))..)))). ( -28.60, z-score = -3.74, R) >droSim1.chr3L_random 96192 91 - 1049610 -UGCUCCCACU-UCCGUUUCCGCAGUUCCCUUGGUUUUAUAUUAUUUC-GGCUGGCUGUAAACGGUGUCUCAUC--GUUUACCGCCAGCUUUAGCC -.(((......-.......(((.((....)))))..............-(((((((.((((((((((...))))--)))))).)))))))..))). ( -27.00, z-score = -3.90, R) >apiMel3.Group2 6018385 90 - 14465785 -UUCUCCCACUCUUUCAUUUUA-GGUUUUCACAUCUUUACAUGUCAU--GAAUGUGUAUAGAAUUUCUUUUAACUAAGUUGCCAACAAUUAUAC-- -.....................-(((.......(((.(((((((...--..))))))).)))....(((......)))..)))...........-- ( -8.30, z-score = 0.27, R) >consensus _UGCUCCCACU_UCCGUUUCCGCUGUUCCCCUGGUUUUAUAUGAUUUC_GGCUGGCUGUAAACGGUGCCUCAUC__GUUUACCGCCAGCUUUAGCC .................................................(((((((.((((((.............)))))).)))))))...... ( -9.13 = -10.35 + 1.23)


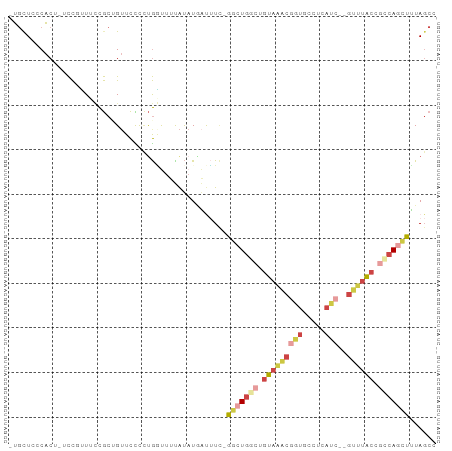
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:56:00 2011