| Sequence ID | dm3.chr3L |
|---|---|
| Location | 1,616,490 – 1,616,591 |
| Length | 101 |
| Max. P | 0.651510 |

| Location | 1,616,490 – 1,616,591 |
|---|---|
| Length | 101 |
| Sequences | 13 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 60.38 |
| Shannon entropy | 0.82532 |
| G+C content | 0.53449 |
| Mean single sequence MFE | -26.76 |
| Consensus MFE | -7.34 |
| Energy contribution | -7.48 |
| Covariance contribution | 0.14 |
| Combinations/Pair | 1.92 |
| Mean z-score | -1.43 |
| Structure conservation index | 0.27 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.34 |
| SVM RNA-class probability | 0.651510 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 1616490 101 + 24543557 ----------UAAUCCCAUUCUGUUUCUUCCUCGCAGGCGCUGCGUGUCCUAUCACGGACGACCGCCCAGCGCCUCGCCAGCGACGGGCAACAAGGUGCAGCACUACAACG- ----------......(((..((((...((((((((((((((((((((((......)))))..)))..))))))).....)))).))).))))..))).............- ( -33.50, z-score = -1.47, R) >droSim1.chr3L 1187360 101 + 22553184 ----------UAAUCCCAUUUUGUUUCUUCCUCGCAGGCGCUGCGUGUCCUACCACGGACGACCGCCCAGCGCCUCGCCAGCGACGGGCAACAAGGUGCAGCACUACAGCG- ----------......(((((((((...((((((((((((((((((((((......)))))..)))..))))))).....)))).))).)))))))))..((......)).- ( -36.70, z-score = -1.95, R) >droSec1.super_2 1642693 101 + 7591821 ----------UAAUCCCAUUUUGUUUCUUCCUCGCAGGCGCUGCGUGUCCUACCACGGACGACCGCCCAGCGCCUCGCCAGCGACGGGCAACAAGGUGCAGCACUACAGCG- ----------......(((((((((...((((((((((((((((((((((......)))))..)))..))))))).....)))).))).)))))))))..((......)).- ( -36.70, z-score = -1.95, R) >droYak2.chr3L 1567647 101 + 24197627 ----------UAAUUCGAUUCUGUUCUUUCAUUGCAGCCGCUGCAUCUCCUACCGCGGACGACCGCCCAGCUCCUCGCCAGCGACGGGCAACAAGGUGCAGCACUACUCCG- ----------..........((((.........))))..(((((((((......((((....))))...((((.(((....))).))))....))))))))).........- ( -32.10, z-score = -2.00, R) >droEre2.scaffold_4784 1595999 99 + 25762168 ----------UAAUCCCAUUCUGUGUUU--CUCGCAGCCGCUGCGUUUCCUAUCACGGACGACCGCCCAGCUCCUCGCCAGCGACGGGCAACAAGGUGCAGCACUACACCG- ----------..........(((((...--..)))))..(((((...(((......)))..(((.....((((.(((....))).)))).....)))))))).........- ( -28.70, z-score = -1.01, R) >droAna3.scaffold_13337 12491305 111 + 23293914 UAGACAAUAAUGUUAACAUAUUCUUUCCCAUCCUUAGACGCUGUGUUUCCUACCAUGGACGACCGCCCAGCUCAUCGCCUGCCACUGGCAACAAGGUGCAGCACAAGACGG- ..((((....)))).............((.((....((.((((.((((((......))).)))....))))))...((.((((..((....))..).)))))....)).))- ( -20.80, z-score = 0.72, R) >dp4.chrXR_group8 4173938 93 + 9212921 ------------------UUUAAUCGCCCAUCCGCAGCCGCUGCAUAUCCUAUAUUGGACGCAAGCCCAACUCCGCUCCGGCCACGGGCAACAAAGUGCAGUUCAAAACAG- ------------------.......((......))....(((((((........((((.(....).))))....((.(((....)))))......))))))).........- ( -23.50, z-score = -1.08, R) >droPer1.super_29 409746 93 + 1099123 ------------------UUUAAACGCCCAUCCACAGCCGCUGCAUAUCCUAUAUUGGACGCAAGCCCAACUCCGCUCCGGCCACGGGCAACAAAGUGCAGUUCAAAACAG- ------------------.....................(((((((........((((.(....).))))....((.(((....)))))......))))))).........- ( -22.60, z-score = -1.40, R) >droWil1.scaffold_180727 1214868 106 + 2741493 -----UAAUUUCUUUACGCCUACUCACUCUUACGCAGCCGGUGUAUAUCCUAUCAUGGACGUCCGCCAAAUUCGGCGCCAUCUACUGGCAAUAAGGUUCGGCAUUAUACUG- -----............(((.....((.((((....(((((((....(((......)))....((((......)))).....)))))))..))))))..))).........- ( -24.10, z-score = -1.19, R) >droVir3.scaffold_13049 19958289 93 - 25233164 ------------------AAACGCAUUCAACCCACAGCCGUGGCAUUAAUUAUCACGGACGACCACCAAACUCGGCCUCAUCCACCGGGAACAAGGUGCAGCAUUAUUCGC- ------------------....((............((((((..(.....)..))))).)...((((...(((((.........))))).....))))..)).........- ( -19.30, z-score = -0.23, R) >droMoj3.scaffold_6680 21124738 93 - 24764193 ------------------AAAAUCAUUUAAUUCACAGCCGCUGUUUCCUUUAUCGUGGACGUCAACCAAAUUCGGCUUCGUCCACGGGCAACAAGUUACAGCAUUUAACCG- ------------------.....................(((((..(......((((((((....((......))...))))))))(....)..)..))))).........- ( -24.70, z-score = -4.13, R) >droGri2.scaffold_15110 13712680 95 + 24565398 ----------------UGAUCUGCUGGCAAUUUACAGGCGCUGCAUUGUUUAUCGUGGACGAUUACCAAAUUCGGCAUCAGCCACUGGCAAUAAAGUACAGCAUUAU-CACG ----------------((((.(((((((.........))(((..((((((....((((..(((..((......)).)))..)))).))))))..))).)))))..))-)).. ( -22.80, z-score = -0.49, R) >apiMel3.Group10 2849481 94 + 11440700 -----------------UAACAACUUUAAUCUUUAACUUUCUGUUUCAGAUAUGUUGGUCGACCAACGGGAGCUUCACCCGCAACAGGAAGUAAAAUGCAGCAUCUCAAAG- -----------------..................((((((((((.(......(((((....)))))(((.......)))).))))))))))...................- ( -22.40, z-score = -2.40, R) >consensus __________U__U__CAUUUUGUUUCUAACUCGCAGCCGCUGCAUUUCCUAUCAUGGACGACCGCCCAGCUCCUCGCCAGCCACGGGCAACAAGGUGCAGCACUACACCG_ .......................................(((((((.(((......))).....................((.....))......))))))).......... ( -7.34 = -7.48 + 0.14)


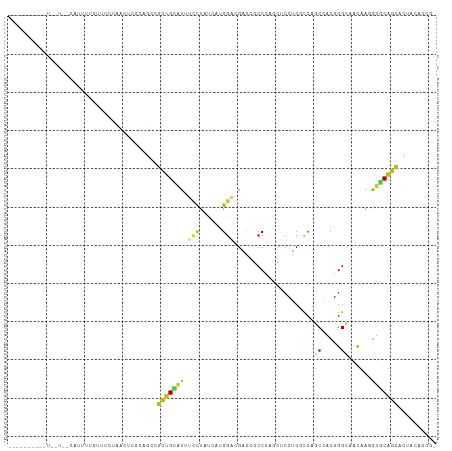
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:55:59 2011