
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 1,603,159 – 1,603,262 |
| Length | 103 |
| Max. P | 0.594550 |

| Location | 1,603,159 – 1,603,262 |
|---|---|
| Length | 103 |
| Sequences | 8 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 64.37 |
| Shannon entropy | 0.65679 |
| G+C content | 0.48634 |
| Mean single sequence MFE | -31.89 |
| Consensus MFE | -13.57 |
| Energy contribution | -11.96 |
| Covariance contribution | -1.61 |
| Combinations/Pair | 1.81 |
| Mean z-score | -0.97 |
| Structure conservation index | 0.43 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.21 |
| SVM RNA-class probability | 0.594550 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 1603159 103 + 24543557 -----------GAAUCGGAUGCCCU-GUCGAGCACUUCGAAUGCCAGGCACAUGGAGUGCCUGUUCGCCAAGUGGCUGGGGGACACUAGCUCGGUCAAUGGAGUGGGUUAUAUGA----- -----------...(((...((((.-.((...(((((((((...(((((((.....)))))))))))..)))))(((((......)))))..........))..))))....)))----- ( -33.80, z-score = 0.36, R) >droSim1.chr3L_random 81698 103 + 1049610 -----------GAAUCGGAUGCUCU-GGCGAGUACUUCGAAUGCCAGGCACAUGGAGUGCCUGUUCGCCACGUGGCUGAGGGACAAUAACUCGGUCAAUGGAGUGGGUUAUGUGA----- -----------(((((((((((((.-...))))).)))))....(((((((.....)))))))))).((((.((((((((.........)))))))).....)))).........----- ( -38.70, z-score = -1.88, R) >droSec1.super_2 1629411 103 + 7591821 -----------GAAUCGGAUGCUCU-GGCGAGUACUUCGAAUGCCAGGCACAUGGAGUGCCUGUUCGCCACGUGGCUGAGGGACAAUAACUCGGUCAAUGGAGUGGGUUAUGUGA----- -----------(((((((((((((.-...))))).)))))....(((((((.....)))))))))).((((.((((((((.........)))))))).....)))).........----- ( -38.70, z-score = -1.88, R) >droYak2.chr3L 1554218 103 + 24197627 -----------GAAUCGGACGUGCU-GGCGAGUACUUCGAAUGCUAGGCACUUGGAGUGCCUGUUCGCCAAGUGGAUGAGGGACAAUAGCUCGGUCAAUGGAGUGGGUGAUAUCA----- -----------...(((..(((..(-((((((((((((((.(((...))).)))))))))....)))))).)))..)))..((.....(((((.((....)).)))))....)).----- ( -31.40, z-score = -0.09, R) >droEre2.scaffold_4784 1582637 103 + 25762168 -----------GAAUCGGAUGUGCU-GGCGAGUGCUUCGAAUGCCAGACACUUGGAGUGCCUGUUCACCAAGUGGAUGAGGGACAAUAACUCGAUCAAUGGAGUGGGUGAUAUAU----- -----------...((.((.(((((-((((..((...))..)))))).))))).))((.(((((((((...)))))).))).))....(((((.((....)).))))).......----- ( -28.70, z-score = -0.27, R) >droAna3.scaffold_13337 12477017 103 + 23293914 -----------AAUUCAGAUGGAUU-CUCGAAUGCCUCGAACUCUAGAUAUAUGGAGUUCCUGUAUGGCAAGUGGAUUCAAGAUGAAAAAUCCAUACAUGAGGUAUGAGGAAAGA----- -----------............((-(((..(((((((((((((((......)))))))).(((((((...((...(((.....)))..))))))))).))))))))))))....----- ( -30.90, z-score = -2.61, R) >dp4.chrXR_group8 4159298 112 + 9212921 CAAAGAGAUGCAAGUGUGACGGCUGAUAUCGGUGCCACCAAUGCCAAAUACGUGGAGCAUUUGUUCUCAAAAUGGCUGGAGGACGAAUCCUCCAUUCAUGUGGUAAGUGUUG-------- ......(((((..(.(((.((.(((....))))).))))..(((((.....((((((.((((((((((..........)))))))))).)))))).....))))).))))).-------- ( -34.10, z-score = -1.39, R) >droWil1.scaffold_180727 1200198 103 + 2741493 ----------------GAAUUGCUU-UGCCUGUGGCACCAAUGCAAAAUAUAUGGAGUACUUAUUUUCAAAAUGGGUGAAAGAUGAAACUUCUGUUCAUGUGGUAUGAAUGAUAAUGUAC ----------------...((((..-((((...)))).....))))..........(((((((((((((...((.((((((((.......))).))))).))...)))).))))).)))) ( -18.80, z-score = 0.01, R) >consensus ___________GAAUCGGAUGCCCU_GGCGAGUACUUCGAAUGCCAGACACAUGGAGUGCCUGUUCGCCAAGUGGCUGAGGGACAAUAACUCGGUCAAUGGAGUGGGUGAUAUGA_____ ................................((((((......(((((((.....)))))))..............(((.........))).......))))))............... (-13.57 = -11.96 + -1.61)

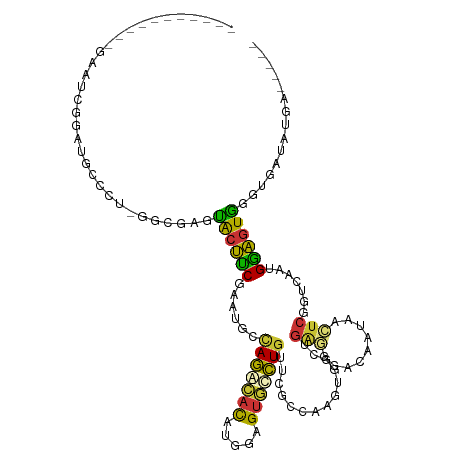
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:55:57 2011