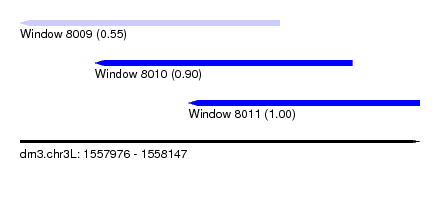
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 1,557,976 – 1,558,147 |
| Length | 171 |
| Max. P | 0.999596 |

| Location | 1,557,976 – 1,558,087 |
|---|---|
| Length | 111 |
| Sequences | 12 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 85.82 |
| Shannon entropy | 0.29032 |
| G+C content | 0.40945 |
| Mean single sequence MFE | -28.14 |
| Consensus MFE | -18.78 |
| Energy contribution | -18.71 |
| Covariance contribution | -0.08 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.71 |
| Structure conservation index | 0.67 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.12 |
| SVM RNA-class probability | 0.551566 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 1557976 111 - 24543557 UUGGAAAUAUUACGCAAGUACUAUUUCUAGUCGUACCCGUUGGAUAAUCAUGGCUCAGUAAACAUGAAUCCGCAACGAGAAUACUUUGCUCAGCGCACUCCGU---GUGGAGAC----- (((((((((.(((....))).))))))))).......((((((((..(((((..........)))))))))((((.(......).))))..))))..((((..---..))))..----- ( -29.60, z-score = -1.60, R) >droSim1.chr3L 1140525 111 - 22553184 UUGGAAAUAUUACGCAAGUACUAUUUCUAGUCGUACCCGUUGGAUAAUCAUGGCUCAGUAAACAUGAAUCCGCAACGAGAAUACUUUGCUCAGCGCACUCCGU---GUGGAGAC----- (((((((((.(((....))).))))))))).......((((((((..(((((..........)))))))))((((.(......).))))..))))..((((..---..))))..----- ( -29.60, z-score = -1.60, R) >droSec1.super_2 1584387 111 - 7591821 UUGGAAAUAUUACGCAAGUACUAUUUCUAGUCGUACCCGUUGGAUAAUCAUGGCUCAGUAAACAUGAAUCCGCAACGAGAAUACUUUGCUCAGCGCACUCCGU---GUGGAGAC----- (((((((((.(((....))).))))))))).......((((((((..(((((..........)))))))))((((.(......).))))..))))..((((..---..))))..----- ( -29.60, z-score = -1.60, R) >droYak2.chr3L 1507280 111 - 24197627 UUGGAAAUAUUACGCAAGUACUAUUUCUAGUCGUACCAGUUGGAUAAUCAUGGCUCAGUAAACAUGAAUCCGCAACGAGAAUACUUUGCUCAGCGCACUCCGU---GUGGAGAC----- .((((((((.(((....))).))))))))(((...((....))....(((((..........))))).((((((..(((.......(((.....))))))..)---))))))))----- ( -29.61, z-score = -1.66, R) >droEre2.scaffold_4784 1536037 111 - 25762168 UUGGAAAUAUUACGCAAGUACUAUUUCUAGUCGUACCCGUUGGUUAAUCAUGGCUCAGUAAACAUGAAUCCGCAACGAGAAUACUUUGCUCAGCGCACUCCGU---GUGGAGAC----- .((((((((.(((....))).))))))))(((..(((....)))...(((((..........))))).((((((..(((.......(((.....))))))..)---))))))))----- ( -29.01, z-score = -1.34, R) >droAna3.scaffold_13337 21394675 110 + 23293914 UUGGAAAUAUUACGCAAGUAUUAUUUCUGGUUGUACC-GUUGGAUAAUCAUGGCUCAGUAAAUAUGAAUCCGCAACGAGAAUACUCUGCCCAACGCACUCCGU---GUGGAGAC----- (..((((((.(((....))).))))))..)......(-(((((........((.(((.......)))..))(((..(((....))))))))))))..((((..---..))))..----- ( -30.40, z-score = -2.03, R) >dp4.chrXR_group6 8160224 111 + 13314419 UUGGAAAUAUUACGCAAGUACUAUUUCUGGUUAUAUCUGUUGAAUAAUCAUGGCUCAGUAAACAUGAAUCCUCAACGAGAAUAUUUUGCCCAACGCACUCCGU---GUGGAGAC----- ..(((((((.(((....))).)))))))(((.(((((((((((....(((((..........)))))....))))).)).))))...))).......((((..---..))))..----- ( -29.30, z-score = -2.63, R) >droPer1.super_9 173581 111 + 3637205 UUGGAAAUAUUACGCAAGUACUAUUUCUGGUUAUAUCUGUUGAAUAAUCAUGGCUCAGUAAACAUGAAUCCUCAACGAGAAUAUUUUGCCCAACGCACUCCGU---GUGGAGAC----- ..(((((((.(((....))).)))))))(((.(((((((((((....(((((..........)))))....))))).)).))))...))).......((((..---..))))..----- ( -29.30, z-score = -2.63, R) >droWil1.scaffold_180916 2625462 114 + 2700594 UUGGAAAUAUUACGAAAGUAUUAUUUCUGGUCAUAUCGUAUUGAUAAUCAUGGCUCAAUAAACAUGAAUCCGCAACGAAAAUCAUUUGCCCAACGCACUCCGCCGAGUGGAGAU----- (..((((((.(((....))).))))))..).......(..(((....(((((..........)))))....((((..........)))).)))..).((((((...))))))..----- ( -28.20, z-score = -2.03, R) >droVir3.scaffold_13049 15134262 115 + 25233164 UUGGAAAUAUUACGCAAGUAUUAUUUCUGGGUAU-AUUUGUUAAUAAUCAUGGCUCAGUAAAGAUGAAUCCUCAACGAAAAAUAAUCGCGCAACGCACUCCGU---GUGGAGACAAACG (..((((((.(((....))).))))))..)....-.((((((......(((((...(((...(.(((....))).)...........(((...))))))))))---)....)))))).. ( -23.50, z-score = -0.76, R) >droMoj3.scaffold_6680 10696142 115 - 24764193 UUGGAAAUAUUACGCAAGUAUUAUUUCUGGGUAU-AUUUGUUAAUAAUCAUGGCUCAGUAAAAAUGAAUCCUCAACGAAAAAUUGUCGCGCAACGCACUCCUU---GUGGAGACGAACG (..((((((.(((....))).))))))..)....-.((((((.....((((............))))......))))))...((((((((...)))..(((..---..))))))))... ( -25.20, z-score = -1.23, R) >droGri2.scaffold_15110 3900662 116 + 24565398 UUGGAAAUAUUACGCAAGUAUUAUUUCUGGAUAUAAUUUGUCAAUAAUCAUGGCCCAGUAAAGAUGAAUCCUCAACGCAAAAUAAUCGCUCAACGCACUCCGU---GUGGAGACAAACG (..((((((.(((....))).))))))..)......((((((......(((((...(((...(.(((....))).)...........((.....)))))))))---)....)))))).. ( -24.40, z-score = -1.43, R) >consensus UUGGAAAUAUUACGCAAGUACUAUUUCUGGUCAUACCUGUUGGAUAAUCAUGGCUCAGUAAACAUGAAUCCGCAACGAGAAUACUUUGCUCAACGCACUCCGU___GUGGAGAC_____ (((((((((.(((....))).))))))))).....................((.(((.......)))..))................((.....)).(((((.....)))))....... (-18.78 = -18.71 + -0.08)
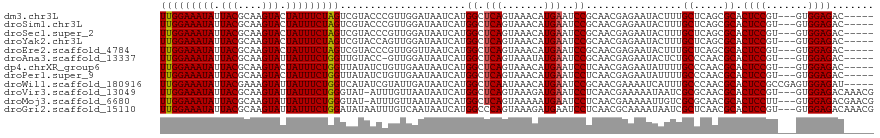
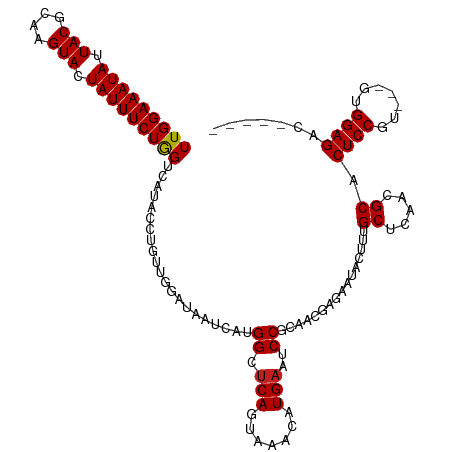
| Location | 1,558,008 – 1,558,118 |
|---|---|
| Length | 110 |
| Sequences | 12 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 77.40 |
| Shannon entropy | 0.48040 |
| G+C content | 0.37253 |
| Mean single sequence MFE | -24.35 |
| Consensus MFE | -12.62 |
| Energy contribution | -12.46 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.85 |
| Structure conservation index | 0.52 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.16 |
| SVM RNA-class probability | 0.902230 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 1558008 110 - 24543557 --GCACUUACUCG--CAAAAGCCGCAUUUGAUUUUUUGGAAAUAUUACGCAAGUACUAUUUCUAGUCGUACCCGUUGGAUAAUCAUGGCUCAGUAAACAUGAAUCCGCAACGAG --.......((((--(....)).((..........(((((((((.(((....))).)))))))))...........((((..(((((..........)))))))))))...))) ( -25.90, z-score = -1.87, R) >droSim1.chr3L 1140557 110 - 22553184 --GCAGUUACUCG--CAAAAGCCGCAUUCGAUUUUUUGGAAAUAUUACGCAAGUACUAUUUCUAGUCGUACCCGUUGGAUAAUCAUGGCUCAGUAAACAUGAAUCCGCAACGAG --.......((((--(....)).((...((((....((((((((.(((....))).))))))))))))........((((..(((((..........)))))))))))...))) ( -27.30, z-score = -2.10, R) >droSec1.super_2 1584419 110 - 7591821 --GCACUUACUCG--CAAAAGCAGCAUUCGAUUUUUUGGAAAUAUUACGCAAGUACUAUUUCUAGUCGUACCCGUUGGAUAAUCAUGGCUCAGUAAACAUGAAUCCGCAACGAG --.......((((--(....)).((...((((....((((((((.(((....))).))))))))))))........((((..(((((..........)))))))))))...))) ( -28.10, z-score = -2.75, R) >droYak2.chr3L 1507312 110 - 24197627 --GCACUUCCUCG--CAAAAGCCGCAUUGGAUUUUUUGGAAAUAUUACGCAAGUACUAUUUCUAGUCGUACCAGUUGGAUAAUCAUGGCUCAGUAAACAUGAAUCCGCAACGAG --.......((((--(....)).(((((((.....(((((((((.(((....))).))))))))).....))))).((((..(((((..........)))))))))))...))) ( -30.20, z-score = -2.97, R) >droEre2.scaffold_4784 1536069 108 - 25762168 ----GCAUCCUCG--CAAAAGCCGCAUUGGAUUUUUUGGAAAUAUUACGCAAGUACUAUUUCUAGUCGUACCCGUUGGUUAAUCAUGGCUCAGUAAACAUGAAUCCGCAACGAG ----.....((((--....(((((.((.((.....(((((((((.(((....))).))))))))).....)).)))))))..(((((..........)))))........)))) ( -26.30, z-score = -1.72, R) >droAna3.scaffold_13337 21394707 110 + 23293914 --ACGCAUACUCAAUCGAAAACUGCAUU-GAAUUUUUGGAAAUAUUACGCAAGUAUUAUUUCUGGUUGUACC-GUUGGAUAAUCAUGGCUCAGUAAAUAUGAAUCCGCAACGAG --..((....(((((((.....)).)))-))((..(..((((((.(((....))).))))))..)..))...-...((((..(((((..........)))))))))))...... ( -23.00, z-score = -0.92, R) >dp4.chrXR_group6 8160256 111 + 13314419 CUGUGCAUUUCUACGUAAGAGUAGCAU---ACCAUUUGGAAAUAUUACGCAAGUACUAUUUCUGGUUAUAUCUGUUGAAUAAUCAUGGCUCAGUAAACAUGAAUCCUCAACGAG .(((((..((((.....))))..))))---)....(..((((((.(((....))).))))))..).......((((((....(((((..........)))))....)))))).. ( -27.20, z-score = -2.36, R) >droPer1.super_9 173613 111 + 3637205 CUGUGCAUUUCUACGUAAGAGUAGCAU---ACCAUUUGGAAAUAUUACGCAAGUACUAUUUCUGGUUAUAUCUGUUGAAUAAUCAUGGCUCAGUAAACAUGAAUCCUCAACGAG .(((((..((((.....))))..))))---)....(..((((((.(((....))).))))))..).......((((((....(((((..........)))))....)))))).. ( -27.20, z-score = -2.36, R) >droWil1.scaffold_180916 2625497 110 + 2700594 -GCAAUUUCCUUG--CAUCAAACUCAAUCAA-UUUUUGGAAAUAUUACGAAAGUAUUAUUUCUGGUCAUAUCGUAUUGAUAAUCAUGGCUCAAUAAACAUGAAUCCGCAACGAA -.....(((.(((--(..........(((((-(..(..((((((.(((....))).))))))..).........))))))..(((((..........)))))....)))).))) ( -24.40, z-score = -3.06, R) >droVir3.scaffold_13049 15134299 106 + 25233164 -----UUAUACCGAAAACCACACGCAU--AACCUUUUGGAAAUAUUACGCAAGUAUUAUUUCUGGGUAU-AUUUGUUAAUAAUCAUGGCUCAGUAAAGAUGAAUCCUCAACGAA -----...(((.((...(((.......--.((((...(((((((.(((....))).))))))))))).(-(((....))))....))).)).)))..(.(((....))).)... ( -17.60, z-score = -0.58, R) >droMoj3.scaffold_6680 10696179 106 - 24764193 -----UUAAACCGAAAACCACUCGCAU--AACCUUUUGGAAAUAUUACGCAAGUAUUAUUUCUGGGUAU-AUUUGUUAAUAAUCAUGGCUCAGUAAAAAUGAAUCCUCAACGAA -----................(((...--.((((...(((((((.(((....))).)))))))))))..-................(..(((.......)))..).....))). ( -17.30, z-score = -0.87, R) >droGri2.scaffold_15110 3900699 107 + 24565398 -----UUAUACCGAAAACGAAUCGCAU--AACCUUUUGGAAAUAUUACGCAAGUAUUAUUUCUGGAUAUAAUUUGUCAAUAAUCAUGGCCCAGUAAAGAUGAAUCCUCAACGCA -----..................((..--........(((((((.(((....))).)))))))((((.......((((.......)))).((.......)).)))).....)). ( -17.70, z-score = -0.65, R) >consensus __GCACUUACUCG__CAAAAGCCGCAUU_GACUUUUUGGAAAUAUUACGCAAGUACUAUUUCUGGUCAUACCUGUUGGAUAAUCAUGGCUCAGUAAACAUGAAUCCGCAACGAG ...................................(((((((((.(((....))).))))))))).....................((.(((.......)))..))........ (-12.62 = -12.46 + -0.16)


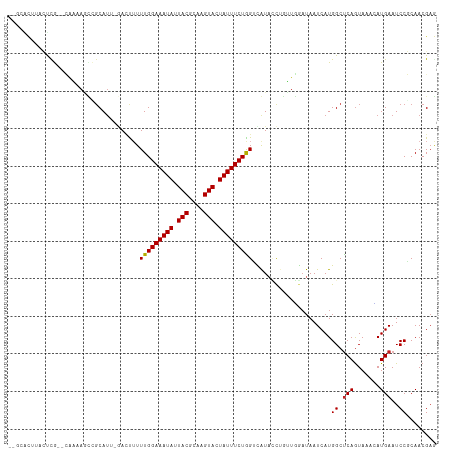
| Location | 1,558,048 – 1,558,147 |
|---|---|
| Length | 99 |
| Sequences | 12 |
| Columns | 107 |
| Reading direction | reverse |
| Mean pairwise identity | 65.24 |
| Shannon entropy | 0.72189 |
| G+C content | 0.35557 |
| Mean single sequence MFE | -20.37 |
| Consensus MFE | -11.68 |
| Energy contribution | -11.44 |
| Covariance contribution | -0.24 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.24 |
| Structure conservation index | 0.57 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 4.06 |
| SVM RNA-class probability | 0.999596 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
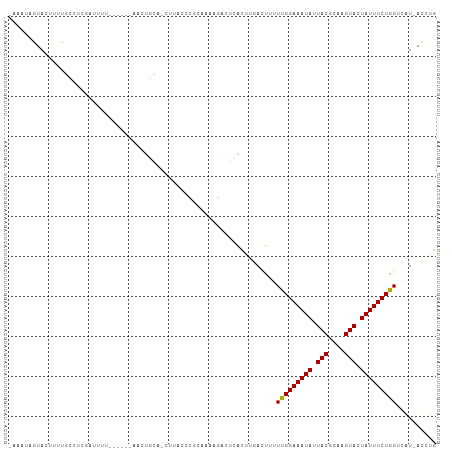
>dm3.chr3L 1558048 99 - 24543557 AAAAAAGUACUUUCGCCUCGAAUUU------AACUGCA-CUUACUCGCAAAAGCCGCAUUUGAUUUUUUGGAAAUAUUACGCAAGUACUAUUUCUAGUCGU-ACCCG ......((((.........(((.((------((.(((.-(((........)))..))).)))).)))(((((((((.(((....))).)))))))))..))-))... ( -19.70, z-score = -2.10, R) >droSim1.chr3L 1140597 98 - 22553184 -AAAAAGUACUUUCGCCUCGAAUUU------AACUGCA-GUUACUCGCAAAAGCCGCAUUCGAUUUUUUGGAAAUAUUACGCAAGUACUAUUUCUAGUCGU-ACCCG -.....((((.......((((((.(------(((....-))))...((.......))))))))....(((((((((.(((....))).)))))))))..))-))... ( -22.00, z-score = -2.46, R) >droSec1.super_2 1584459 98 - 7591821 -AAAAAGUACUUUCGCCUCGAAUUU------AACUGCA-CUUACUCGCAAAAGCAGCAUUCGAUUUUUUGGAAAUAUUACGCAAGUACUAUUUCUAGUCGU-ACCCG -.....((((.......((((((..------..((((.-.............)))).))))))....(((((((((.(((....))).)))))))))..))-))... ( -23.24, z-score = -3.38, R) >droYak2.chr3L 1507352 98 - 24197627 -CAAAAGUACUUUCGCCUCGAUUUU------AACUGCA-CUUCCUCGCAAAAGCCGCAUUGGAUUUUUUGGAAAUAUUACGCAAGUACUAUUUCUAGUCGU-ACCAG -.....((((...............------.......-..(((..((.......))...)))....(((((((((.(((....))).)))))))))..))-))... ( -19.00, z-score = -1.44, R) >droEre2.scaffold_4784 1536109 96 - 25762168 -AAAAAGUACUUUCGCCUCGAUUUU------AACCGCA---UCCUCGCAAAAGCCGCAUUGGAUUUUUUGGAAAUAUUACGCAAGUACUAUUUCUAGUCGU-ACCCG -.....((((...............------......(---(((..((.......))...))))...(((((((((.(((....))).)))))))))..))-))... ( -19.70, z-score = -1.75, R) >droAna3.scaffold_13337 21394747 104 + 23293914 -AUAUAGUACUUUAGCCACCGACUCUUCGGGCUCUACGCAUACUCAAUCGAAAACUGCAUUGAAUUUUUGGAAAUAUUACGCAAGUAUUAUUUCUGGUUGU-ACCG- -.....((((...((((.((((....))))((.....))....(((((((.....)).)))))......(((((((.(((....))).)))))))))))))-))..- ( -23.90, z-score = -2.47, R) >dp4.chrXR_group6 8160296 97 + 13314419 ---UUAGUACUUUUAAAUCGAUUUA-----CGCUCUGUGCAUUUCUACGUAAGAGUAGCAU-ACCAUUUGGAAAUAUUACGCAAGUACUAUUUCUGGUUAU-AUCUG ---....((((((((....((..(.-----(((...))).)..))....))))))))....-.....(..((((((.(((....))).))))))..)....-..... ( -19.00, z-score = -1.33, R) >droPer1.super_9 173653 97 + 3637205 ---UUAGUACUUUUAAAUCGAUUUA-----CGCUCUGUGCAUUUCUACGUAAGAGUAGCAU-ACCAUUUGGAAAUAUUACGCAAGUACUAUUUCUGGUUAU-AUCUG ---....((((((((....((..(.-----(((...))).)..))....))))))))....-.....(..((((((.(((....))).))))))..)....-..... ( -19.00, z-score = -1.33, R) >droWil1.scaffold_180916 2625537 98 + 2700594 -AUCUAGUACAUUUUACUUUGAUUU------GCUUGCAAUUUCCUUGCAUCAAACUCAAUCAA-UUUUUGGAAAUAUUACGAAAGUAUUAUUUCUGGUCAU-AUCGU -.................(((((((------(..(((((.....))))).)))).))))....-...(..((((((.(((....))).))))))..)....-..... ( -20.30, z-score = -3.34, R) >droVir3.scaffold_13049 15134338 99 + 25233164 -AUCUAGUACUUUUCACCGC-UUCG-----AUUUUCGAUUUAUACCGAAAACCACACGCAUAACCUUUUGGAAAUAUUACGCAAGUAUUAUUUCUGGGUAU-AUUUG -.................((-...(-----.((((((........)))))).)....))...((((...(((((((.(((....))).)))))))))))..-..... ( -19.50, z-score = -2.72, R) >droMoj3.scaffold_6680 10696218 100 - 24764193 -AUCUAGUACUUUUUAACGCUUUCG-----AUUAUCGACUUAAACCGAAAACCACUCGCAUAACCUUUUGGAAAUAUUACGCAAGUAUUAUUUCUGGGUAU-AUUUG -.................((..(((-----.....)))........((.......))))...((((...(((((((.(((....))).)))))))))))..-..... ( -17.30, z-score = -1.91, R) >droGri2.scaffold_15110 3900738 100 + 24565398 -UUGUGGUACUUUUCGACGC-UUCA-----AUUUUCGAUUUAUACCGAAAACGAAUCGCAUAACCUUUUGGAAAUAUUACGCAAGUAUUAUUUCUGGAUAUAAUUUG -.((((((.(((((((....-.((.-----......)).......)))))).).))))))......((..((((((.(((....))).))))))..))......... ( -21.80, z-score = -2.62, R) >consensus _AAAUAGUACUUUUGCCUCGAUUUU______AACUGCA_CUUACCCGCAAAAGACUCACUUGACUUUUUGGAAAUAUUACGCAAGUACUAUUUCUGGUCAU_ACCUG ...................................................................(((((((((.(((....))).))))))))).......... (-11.68 = -11.44 + -0.24)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:55:53 2011