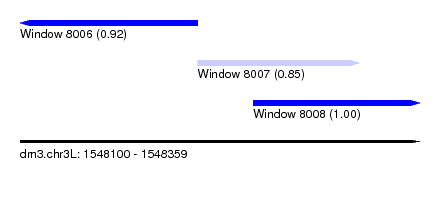
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 1,548,100 – 1,548,359 |
| Length | 259 |
| Max. P | 0.995989 |

| Location | 1,548,100 – 1,548,215 |
|---|---|
| Length | 115 |
| Sequences | 6 |
| Columns | 121 |
| Reading direction | reverse |
| Mean pairwise identity | 75.26 |
| Shannon entropy | 0.46219 |
| G+C content | 0.51189 |
| Mean single sequence MFE | -33.58 |
| Consensus MFE | -18.48 |
| Energy contribution | -20.28 |
| Covariance contribution | 1.81 |
| Combinations/Pair | 1.24 |
| Mean z-score | -1.88 |
| Structure conservation index | 0.55 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.27 |
| SVM RNA-class probability | 0.919154 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
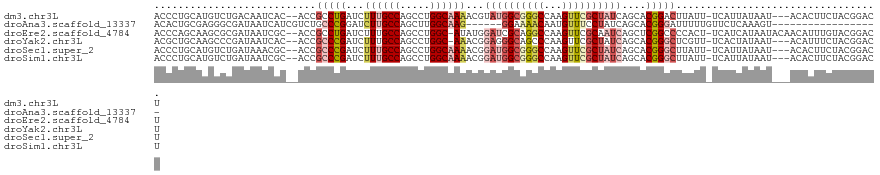
>dm3.chr3L 1548100 115 - 24543557 ACCCUGCAUGUCUGACAAUCAC--ACCGCCUGAUCUUUGCCAGCCUGGCAAAACGUAUGGCGGGCCAAGUUCGCUAUCAGCACGGACUUAUU-UCAUUAUAAU---ACACUUCUACGGACU .........(((((........--.(((((((...((((((.....))))))...)).)))))...(((((((.........)))))))...-..........---.........))))). ( -29.90, z-score = -1.72, R) >droAna3.scaffold_13337 21385607 97 + 23293914 ACACUGCGAGGGCGAUAAUCAUCGUCUGCCCGGAUCUUGCCAGCUUGGCAAG------GGAAAACAAUGUUUCCUAUCAGCACGGGAUUUUUGUUCUCAAAGU------------------ ..(((..((((((((.((((.(((((((.......((((((.....))))))------(((((......)))))...))).)))))))).))))))))..)))------------------ ( -33.50, z-score = -2.63, R) >droEre2.scaffold_4784 1526228 117 - 25762168 ACCCAGCAAGCGCGAUAAUCGC--ACCGCCUGAUCUUUGCCAGCCUGGC-AUAUGGAUCGCAGGCCAAGUUCGCAAUCAGCUCGGCCCCACU-UCAUCAUAAUACAACAUUUGUACGGACU .((..(((((((((.....)))--.......(((((.((((.....)))-)...))))))).((((.((((.......)))).)))).....-.................))))..))... ( -31.80, z-score = -1.02, R) >droYak2.chr3L 1497243 114 - 24197627 ACGCUGCAAGCCCGAUAAUCAC--ACCGCCCGAUCUUUGCCAGCCUGGC-AAACGGAGGGCAGCCCAAGUUCGCUAUCAGCACGGGCUCGUU-UCACUAUAAU---ACAUUUCUACGGACU ..(..((.((((((........--...((((..(.((((((.....)))-))).)..))))...........((.....)).)))))).)).-.)........---.....((....)).. ( -33.90, z-score = -1.49, R) >droSec1.super_2 1574424 115 - 7591821 ACCCUGCAUGUCUGAUAAACGC--ACCGCCCGAUCUUUGCCAGCCUGGCAAAACGGAUGGCGGGCCAAGUUCGCUAUCAGCACGGGCUUAUU-UCAUUAUAAU---ACACUUCUACGGACU .........(((((........--...(((((...((((((.....))))))...((((((((((...))))))))))....)))))((((.-.....)))).---.........))))). ( -36.20, z-score = -2.27, R) >droSim1.chr3L 1130303 115 - 22553184 ACCCUGCAUGUCUGAUAAUCGC--ACCGCCCGAUCUUUGCCAGCCUGGCAAAACGGAUGGCGGGCCAAGUUCGCUAUCAGCACGGGCUUAUU-UCAUUAUAAU---ACACUUCUACGGACU .........(((((........--...(((((...((((((.....))))))...((((((((((...))))))))))....)))))((((.-.....)))).---.........))))). ( -36.20, z-score = -2.16, R) >consensus ACCCUGCAAGUCCGAUAAUCAC__ACCGCCCGAUCUUUGCCAGCCUGGCAAAACGGAUGGCAGGCCAAGUUCGCUAUCAGCACGGGCUUAUU_UCAUUAUAAU___ACACUUCUACGGACU ...........................(((((...((((((.....))))))...((((((((((...))))))))))....))))).................................. (-18.48 = -20.28 + 1.81)

| Location | 1,548,215 – 1,548,320 |
|---|---|
| Length | 105 |
| Sequences | 5 |
| Columns | 109 |
| Reading direction | forward |
| Mean pairwise identity | 92.68 |
| Shannon entropy | 0.12402 |
| G+C content | 0.43678 |
| Mean single sequence MFE | -28.20 |
| Consensus MFE | -24.64 |
| Energy contribution | -23.44 |
| Covariance contribution | -1.20 |
| Combinations/Pair | 1.19 |
| Mean z-score | -1.92 |
| Structure conservation index | 0.87 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.91 |
| SVM RNA-class probability | 0.850097 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 1548215 105 + 24543557 GUUUGUCGAGUGCUAACCAUAAUUAAAGGCGAAUUU----GUCUUAUCGCUGGAAACAGUGAAACAUGGCUCGUUUCCGCCAAACGCAUUUAUCGUGUUUCCAGAUACC ....(..((((((............((((((....)----))))).((((((....))))))....((((........))))...))))))..)(((((....))))). ( -27.60, z-score = -1.98, R) >droEre2.scaffold_4784 1526345 105 + 25762168 GUUUGCUGAGUGCUAACCAUAAUUAAAGGUAAAUGU----GUCUUAUCGCUGGAAACAGUGAAACACGGCUCGUUUCCGCCAAACGCAUUUAUCGUGUUUCCAGAUACU (((.((.....)).)))..........(((((((((----((....((((((....)))))).....(((........)))..)))))))))))(((((....))))). ( -32.90, z-score = -3.67, R) >droYak2.chr3L 1497357 109 + 24197627 GUUUGCUAAGUGCUAACCAUAAUUAAAGGUGAAUUUACCAGUCUUAUCGCUGGUAACAGUGAAACAUGGCCCGUUUCCGCCAAGCGCAUUUAUCGUGUUUCCAGAUACU (((.((.....)).)))..........(((((((((((((((......))))))))..(((.....((((........))))..))))))))))(((((....))))). ( -25.30, z-score = -1.13, R) >droSec1.super_2 1574539 105 + 7591821 GUUUGCCGAGUGCUAACCACAAUUAAAGGCGAAUUG----GUCUUAUCGCUGGAAACAGUGAAACAUGGCUCGUUUCCGCCAAACGCAUUUAUCGUGUUUCCAGAUACC .......((((((............(((((......----))))).((((((....))))))....((((........))))...))))))...(((((....))))). ( -27.60, z-score = -1.38, R) >droSim1.chr3L 1130418 105 + 22553184 GUUUGCCGAGUGCUAACCAUAAUUAAAGGCGAAUUG----GUCUUAUCGCUGGAAACAGUGAAACACGGCUCGUUUCCGCCAAACGCAUUUAUCGUGUUUCCAGAUACC (((((((.(((..........)))...)))))))..----....((((((((....))))(((((((((..(((((.....)))))......)))))))))..)))).. ( -27.60, z-score = -1.42, R) >consensus GUUUGCCGAGUGCUAACCAUAAUUAAAGGCGAAUUU____GUCUUAUCGCUGGAAACAGUGAAACAUGGCUCGUUUCCGCCAAACGCAUUUAUCGUGUUUCCAGAUACC (((((((.(((..........)))...)))))))..........((((((((....))))(((((((((..(((((.....)))))......)))))))))..)))).. (-24.64 = -23.44 + -1.20)

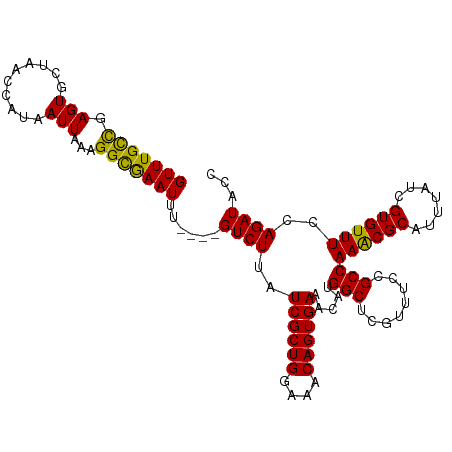
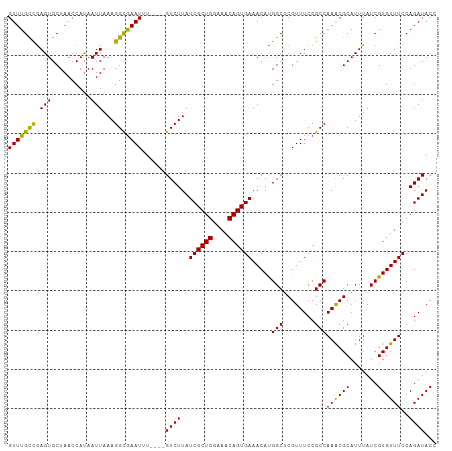
| Location | 1,548,251 – 1,548,359 |
|---|---|
| Length | 108 |
| Sequences | 6 |
| Columns | 109 |
| Reading direction | forward |
| Mean pairwise identity | 80.00 |
| Shannon entropy | 0.38688 |
| G+C content | 0.51316 |
| Mean single sequence MFE | -38.22 |
| Consensus MFE | -22.11 |
| Energy contribution | -24.03 |
| Covariance contribution | 1.92 |
| Combinations/Pair | 1.26 |
| Mean z-score | -3.12 |
| Structure conservation index | 0.58 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.87 |
| SVM RNA-class probability | 0.995989 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
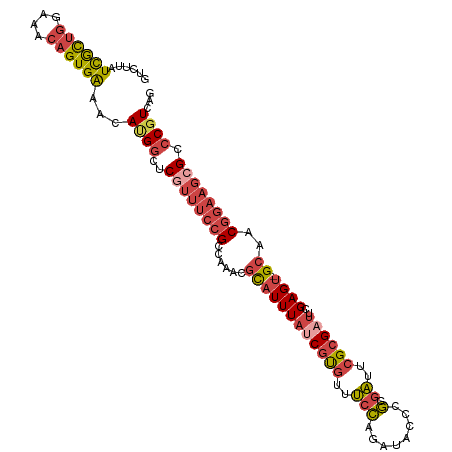
>dm3.chr3L 1548251 108 + 24543557 GUCUUAUCGCUGGAAACAGUGAAACAUGGCUCGUUUCCGCCAAACGCAUUUAUCGUGUUUCCAGAUACCCGCGAUUCGCGAUUCGAGU-GCAGCGGAAGCGGCCGUCAG ......((((((....))))))...(((((.(((((((((.....((((((((((((..(((........).))..))))))..))))-)).))))))))))))))... ( -46.30, z-score = -4.73, R) >droAna3.scaffold_13337 21385737 102 - 23293914 -UUUUCUUAUCUUUAUCAGUGGUGUA-GAAACAACAAGGCCAU---UUUUUUCCGCCAAACUAGAAACACAAAUAUCGGGUUUCGAGUUGCAACGGAAGCGCGCCUG-- -.(((((((((.........)))).)-)))).....((((...---...((((((...((((.(((((.(........)))))).))))....))))))...)))).-- ( -21.30, z-score = -0.36, R) >droEre2.scaffold_4784 1526381 108 + 25762168 GUCUUAUCGCUGGAAACAGUGAAACACGGCUCGUUUCCGCCAAACGCAUUUAUCGUGUUUCCAGAUACUCGCGAUUCGCGUUUCGAGU-GCAACGGAAGAGCCCGUCAC ......((((((....)))))).....(((((...((((..((((((.......))))))...(.((((((.(((....))).)))))-))..)))).)))))...... ( -35.50, z-score = -2.12, R) >droYak2.chr3L 1497397 108 + 24197627 GUCUUAUCGCUGGUAACAGUGAAACAUGGCCCGUUUCCGCCAAGCGCAUUUAUCGUGUUUCCAGAUACUCGCGAUUCGCGAUUCGAGU-GCAACGGAAGCGCCCGUCAG ......((((((....))))))....((((.((((((((....((((....((((((..(((........).))..))))))....))-))..))))))))...)))). ( -36.00, z-score = -2.08, R) >droSec1.super_2 1574575 108 + 7591821 GUCUUAUCGCUGGAAACAGUGAAACAUGGCUCGUUUCCGCCAAACGCAUUUAUCGUGUUUCCAGAUACCCGCGAUUCGCGAUUCGAGU-GCAGCGGAAGCGACCGUCAG ......((((((....))))))...((((.((((((((((.....((((((((((((..(((........).))..))))))..))))-)).))))))))))))))... ( -44.20, z-score = -4.50, R) >droSim1.chr3L 1130454 108 + 22553184 GUCUUAUCGCUGGAAACAGUGAAACACGGCUCGUUUCCGCCAAACGCAUUUAUCGUGUUUCCAGAUACCCGCGAUUCGCGAUUCGAGU-GCAGCGGAAGCGACCGUCAG ......((((((....))))))...((((.((((((((((.....((((((((((((..(((........).))..))))))..))))-)).))))))))))))))... ( -46.00, z-score = -4.92, R) >consensus GUCUUAUCGCUGGAAACAGUGAAACAUGGCUCGUUUCCGCCAAACGCAUUUAUCGUGUUUCCAGAUACCCGCGAUUCGCGAUUCGAGU_GCAACGGAAGCGCCCGUCAG ......((((((....))))))...((((..((((((((..((((((.......))))))..........((.(((((.....))))).))..)))))))).))))... (-22.11 = -24.03 + 1.92)
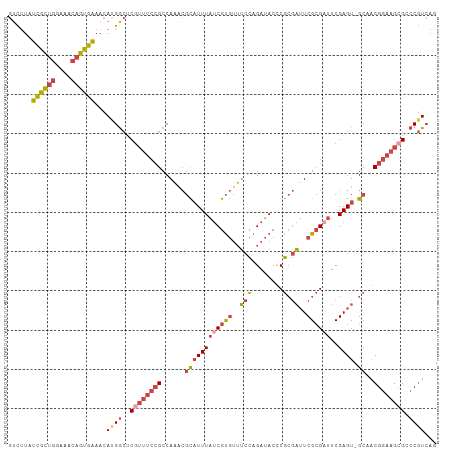
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:55:51 2011