| Sequence ID | dm3.chr2L |
|---|---|
| Location | 4,128,556 – 4,128,650 |
| Length | 94 |
| Max. P | 0.998718 |

| Location | 4,128,556 – 4,128,650 |
|---|---|
| Length | 94 |
| Sequences | 5 |
| Columns | 97 |
| Reading direction | forward |
| Mean pairwise identity | 92.21 |
| Shannon entropy | 0.13272 |
| G+C content | 0.59283 |
| Mean single sequence MFE | -24.00 |
| Consensus MFE | -18.65 |
| Energy contribution | -20.05 |
| Covariance contribution | 1.40 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.26 |
| Structure conservation index | 0.78 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.75 |
| SVM RNA-class probability | 0.806908 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
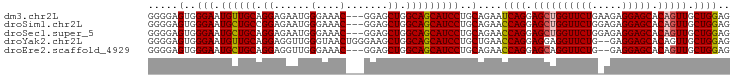
>dm3.chr2L 4128556 94 + 23011544 CUCCAGCAACUGUGCUCCUCUUCAGAACCAGCUCCUGAUUCUGCAGGAUGCUGCCAGCUCC---GUUUCCCAUUCUCCUGCAACAUUCCCACUCCCC .....(((.(((..........)))...(((((((((......))))).))))........---..............)))................ ( -16.40, z-score = -0.45, R) >droSim1.chr2L 4084016 94 + 22036055 CUCCAGCAACUGUGCUCCUCUCCAGAACCAGCUCCUGGUUCUGCAGGAUGCUGCCAGCUCC---GUUUCCCAUUCUCCGGCAGCAUUCCCACUCCCC ....((((....))))......(((((((((...)))))))))..((((((((((......---..............))))))))))......... ( -29.95, z-score = -3.43, R) >droSec1.super_5 2227213 94 + 5866729 CUCCAGCAACUGUGCUCCUCUCCAGAACCAGCUCCUGGUUCUGCAGGAUGCUGCCAGCUCC---GUUUCCCAUUCUCCUGCAGCAUUCCCACUCCCC ....((((....))))......(((((((((...)))))))))..(((((((((.((....---.............)))))))))))......... ( -26.43, z-score = -2.66, R) >droYak2.chr2L 4150295 95 + 22324452 CUCCAGCAACUGUGCUCCUC--CAGAACCUCCUCCUGGUUCAGCAGGAUGCUGCCAGCUUCCCAGUUACCCAACCUCCUGCAACAUUCCCACUCCCC ....((((....))))....--..(((((.......))))).((((((.(.((..((((....))))...)).).))))))................ ( -20.80, z-score = -1.91, R) >droEre2.scaffold_4929 4188832 92 + 26641161 CUCCAGCAACUGUGCUCCUC--CAGAACCUGCUCCUGGUUCUGCAGGAUGCUGCCAGCUCC---GUUUCCCAACCUCCUGCAGCAUUCCCACUCCCC .....(((....)))((((.--(((((((.......))))))).))))((((((.((....---(((....)))...))))))))............ ( -26.40, z-score = -2.85, R) >consensus CUCCAGCAACUGUGCUCCUCU_CAGAACCAGCUCCUGGUUCUGCAGGAUGCUGCCAGCUCC___GUUUCCCAUUCUCCUGCAGCAUUCCCACUCCCC ....((((....))))......(((((((((...)))))))))..(((((((((.........................)))))))))......... (-18.65 = -20.05 + 1.40)
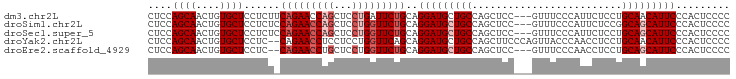
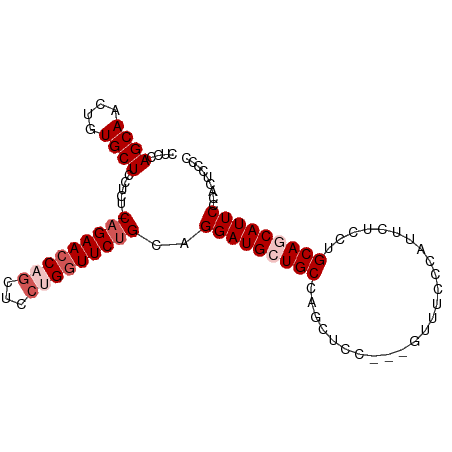
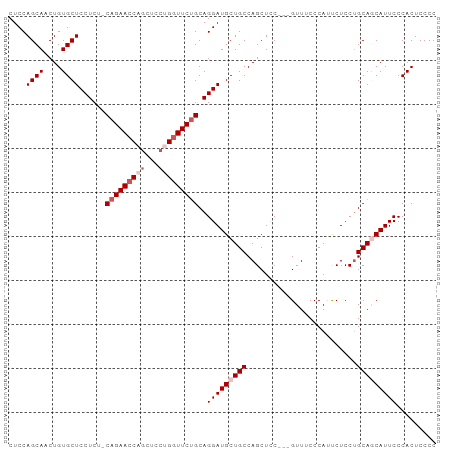
| Location | 4,128,556 – 4,128,650 |
|---|---|
| Length | 94 |
| Sequences | 5 |
| Columns | 97 |
| Reading direction | reverse |
| Mean pairwise identity | 92.21 |
| Shannon entropy | 0.13272 |
| G+C content | 0.59283 |
| Mean single sequence MFE | -39.06 |
| Consensus MFE | -36.12 |
| Energy contribution | -36.32 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.07 |
| Mean z-score | -4.03 |
| Structure conservation index | 0.92 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.46 |
| SVM RNA-class probability | 0.998718 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
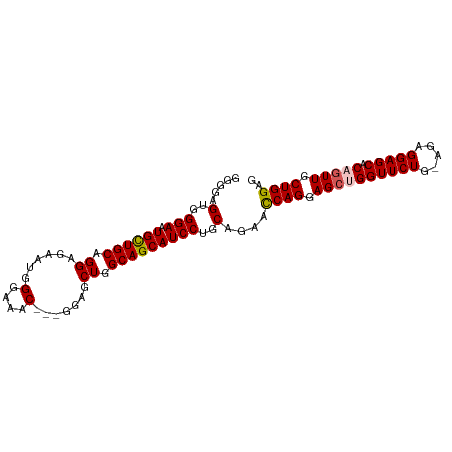
>dm3.chr2L 4128556 94 - 23011544 GGGGAGUGGGAAUGUUGCAGGAGAAUGGGAAAC---GGAGCUGGCAGCAUCCUGCAGAAUCAGGAGCUGGUUCUGAAGAGGAGCACAGUUGCUGGAG .....(..(((.((((((...((..(.(....)---.)..)).)))))))))..)....((((.((((((((((.....))))).))))).)))).. ( -36.80, z-score = -4.39, R) >droSim1.chr2L 4084016 94 - 22036055 GGGGAGUGGGAAUGCUGCCGGAGAAUGGGAAAC---GGAGCUGGCAGCAUCCUGCAGAACCAGGAGCUGGUUCUGGAGAGGAGCACAGUUGCUGGAG .....(..(((.(((((((((....(.(....)---.)..))))))))))))..)....((((.((((((((((.....))))).))))).)))).. ( -47.10, z-score = -6.00, R) >droSec1.super_5 2227213 94 - 5866729 GGGGAGUGGGAAUGCUGCAGGAGAAUGGGAAAC---GGAGCUGGCAGCAUCCUGCAGAACCAGGAGCUGGUUCUGGAGAGGAGCACAGUUGCUGGAG .....(..(((.((((((...((..(.(....)---.)..)).)))))))))..)....((((.((((((((((.....))))).))))).)))).. ( -42.00, z-score = -4.95, R) >droYak2.chr2L 4150295 95 - 22324452 GGGGAGUGGGAAUGUUGCAGGAGGUUGGGUAACUGGGAAGCUGGCAGCAUCCUGCUGAACCAGGAGGAGGUUCUG--GAGGAGCACAGUUGCUGGAG ....((..(((.((((((.((...(..(....)..)....)).)))))))))..))...((((.((..(((((..--...)))).)..)).)))).. ( -30.40, z-score = -1.00, R) >droEre2.scaffold_4929 4188832 92 - 26641161 GGGGAGUGGGAAUGCUGCAGGAGGUUGGGAAAC---GGAGCUGGCAGCAUCCUGCAGAACCAGGAGCAGGUUCUG--GAGGAGCACAGUUGCUGGAG .....(..(((.((((((...((.((.(....)---.)).)).)))))))))..)....((((.(((..((((..--...))))...))).)))).. ( -39.00, z-score = -3.82, R) >consensus GGGGAGUGGGAAUGCUGCAGGAGAAUGGGAAAC___GGAGCUGGCAGCAUCCUGCAGAACCAGGAGCUGGUUCUG_AGAGGAGCACAGUUGCUGGAG .....(..(((.((((((.((......(....).......)).)))))))))..)....((((.((((((((((.....))))).))))).)))).. (-36.12 = -36.32 + 0.20)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:15:26 2011