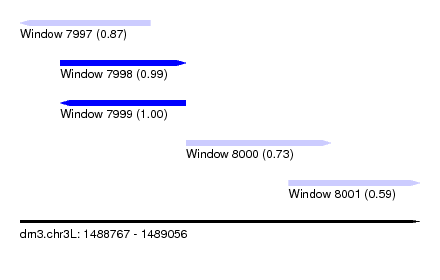
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 1,488,767 – 1,489,056 |
| Length | 289 |
| Max. P | 0.996978 |

| Location | 1,488,767 – 1,488,861 |
|---|---|
| Length | 94 |
| Sequences | 4 |
| Columns | 94 |
| Reading direction | reverse |
| Mean pairwise identity | 73.28 |
| Shannon entropy | 0.42110 |
| G+C content | 0.50265 |
| Mean single sequence MFE | -21.93 |
| Consensus MFE | -13.05 |
| Energy contribution | -13.43 |
| Covariance contribution | 0.37 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.77 |
| Structure conservation index | 0.60 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.01 |
| SVM RNA-class probability | 0.872675 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
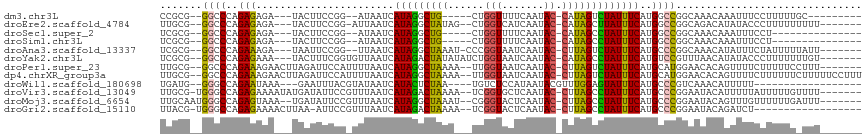
>dm3.chr3L 1488767 94 - 24543557 ACGAAAUGAAUGAAUUAUUUUUUUUAUUCAUCUCGACACAACCUCGGCGGCACUCGAACCGCCACUCGAAAAACGUGAGCACGCGUGAAUCGAC .(((.(((((((((........))))))))).)))..........(((((........)))))..((((...(((((....)))))...)))). ( -29.90, z-score = -4.13, R) >droEre2.scaffold_4784 1473390 71 - 25762168 -----------------------AAAUCACUUUCAUUCCGACCUCGGCGGCACUCGAACCGCCACUCGAAAAUCAUGAGCGCGCGAGAAUCAAC -----------------------............(((((.(((((((((........)))))....(.....)..))).))).)))....... ( -14.60, z-score = -0.32, R) >droSec1.super_2 1523408 84 - 7591821 ACGAAAUGAAUGGAAU----------UUCAUUUCACCCCGACCUCAGCGGCACUCGAACCGCCACUCGAAAAUCAUGAGCGCGUGAGAAUCGAC ..((((((((......----------))))))))............((((........))))...((((...(((((....)))))...)))). ( -22.00, z-score = -1.68, R) >droSim1.chr3L 1078799 83 - 22553184 ACGAAAUGAAUGAAU-----------UUCAUAUCAUCCCGACCUCGGCGGCACUCGAACCGCCACUCGAAAAUCAUGAGCGCGUGAGAAUGGAC ..((.(((((.....-----------))))).))...(((..((((((((........))))).((((.......)))).....)))..))).. ( -21.20, z-score = -0.97, R) >consensus ACGAAAUGAAUGAAU___________UUCAUUUCAUCCCGACCUCGGCGGCACUCGAACCGCCACUCGAAAAUCAUGAGCGCGCGAGAAUCGAC .............................................(((((........)))))..((((...(((((....)))))...)))). (-13.05 = -13.43 + 0.37)

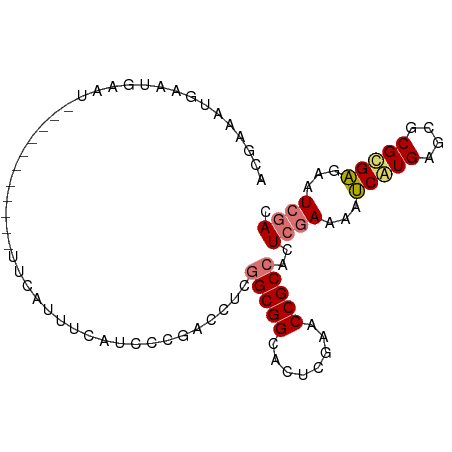
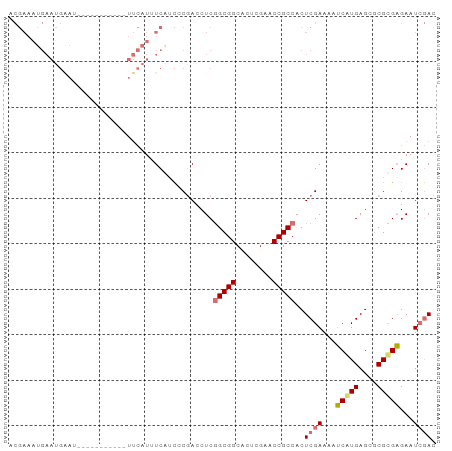
| Location | 1,488,796 – 1,488,887 |
|---|---|
| Length | 91 |
| Sequences | 3 |
| Columns | 92 |
| Reading direction | forward |
| Mean pairwise identity | 73.68 |
| Shannon entropy | 0.35386 |
| G+C content | 0.38581 |
| Mean single sequence MFE | -23.17 |
| Consensus MFE | -18.35 |
| Energy contribution | -18.36 |
| Covariance contribution | 0.01 |
| Combinations/Pair | 1.19 |
| Mean z-score | -2.46 |
| Structure conservation index | 0.79 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.52 |
| SVM RNA-class probability | 0.992192 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 1488796 91 + 24543557 GUGGCGGUUCGAGUGCCGCCGAGGUUGUGUCGAGAUGAAUAAAAAAAAUAAUUCAUUCAUUUCGUUUUCUUAAGAAA-AGAAAUUAAGAUUU .(((((((......))))))).........((((((((((.((........)).))))))))))...((((((....-.....))))))... ( -23.70, z-score = -2.64, R) >droSec1.super_2 1523437 78 + 7591821 GUGGCGGUUCGAGUGCCGCUGAGGUCGGGGUGAAAUGAA----------AUUCCAUUCAUUUCGUUUUCUUAGUAAAUGUAUAAAGAU---- ..((((.......))))(((((((.((..(((((.((..----------....)))))))..))...)))))))..............---- ( -21.40, z-score = -2.01, R) >droSim1.chr3L 1078828 81 + 22553184 GUGGCGGUUCGAGUGCCGCCGAGGUCGGGAUGAUAUGAA----------AUUC-AUUCAUUUCGUUUUCUUAAUAAAUAUAUGUAGAUAUUU ..((((((......))))))((((.((..((((.(((..----------...)-))))))..)).)))).....((((((......)))))) ( -24.40, z-score = -2.71, R) >consensus GUGGCGGUUCGAGUGCCGCCGAGGUCGGGAUGAAAUGAA__________AUUCCAUUCAUUUCGUUUUCUUAAUAAAUAUAUAUAGAUAUUU ..((((((......))))))((((....(((((((((((................))))))))))).))))..................... (-18.35 = -18.36 + 0.01)

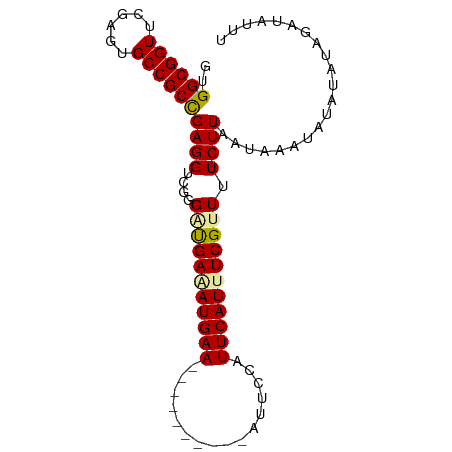
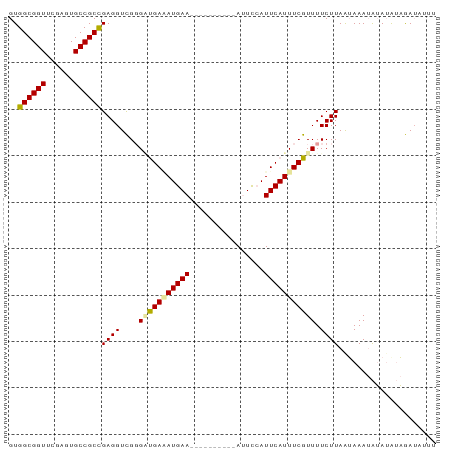
| Location | 1,488,796 – 1,488,887 |
|---|---|
| Length | 91 |
| Sequences | 3 |
| Columns | 92 |
| Reading direction | reverse |
| Mean pairwise identity | 73.68 |
| Shannon entropy | 0.35386 |
| G+C content | 0.38581 |
| Mean single sequence MFE | -17.70 |
| Consensus MFE | -9.92 |
| Energy contribution | -10.26 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -3.48 |
| Structure conservation index | 0.56 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.02 |
| SVM RNA-class probability | 0.996978 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 1488796 91 - 24543557 AAAUCUUAAUUUCU-UUUCUUAAGAAAACGAAAUGAAUGAAUUAUUUUUUUUAUUCAUCUCGACACAACCUCGGCGGCACUCGAACCGCCAC .........(((((-(.....)))))).(((.(((((((((........))))))))).)))..........(((((........))))).. ( -23.90, z-score = -5.58, R) >droSec1.super_2 1523437 78 - 7591821 ----AUCUUUAUACAUUUACUAAGAAAACGAAAUGAAUGGAAU----------UUCAUUUCACCCCGACCUCAGCGGCACUCGAACCGCCAC ----.((((..((....))..))))....((((((((......----------))))))))............((((........))))... ( -14.20, z-score = -2.56, R) >droSim1.chr3L 1078828 81 - 22553184 AAAUAUCUACAUAUAUUUAUUAAGAAAACGAAAUGAAU-GAAU----------UUCAUAUCAUCCCGACCUCGGCGGCACUCGAACCGCCAC ((((((......))))))..........((..((((((-(...----------..))).))))..)).....(((((........))))).. ( -15.00, z-score = -2.30, R) >consensus AAAUAUCUAUAUAUAUUUAUUAAGAAAACGAAAUGAAUGGAAU__________UUCAUAUCAACCCGACCUCGGCGGCACUCGAACCGCCAC .............................((.(((((................))))).))...........(((((........))))).. ( -9.92 = -10.26 + 0.33)

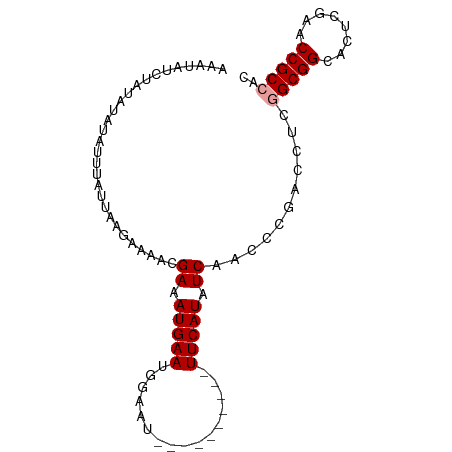
| Location | 1,488,887 – 1,488,992 |
|---|---|
| Length | 105 |
| Sequences | 12 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 66.44 |
| Shannon entropy | 0.64849 |
| G+C content | 0.42723 |
| Mean single sequence MFE | -27.54 |
| Consensus MFE | -10.73 |
| Energy contribution | -9.84 |
| Covariance contribution | -0.89 |
| Combinations/Pair | 1.67 |
| Mean z-score | -1.38 |
| Structure conservation index | 0.39 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.54 |
| SVM RNA-class probability | 0.734578 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
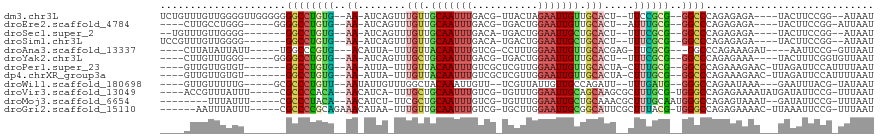
>dm3.chr3L 1488887 105 + 24543557 UCUGUUUGUUGGGGUUGGGGGGGCCUGUG--AA-AUCAGUUUGUUGCAAUUUGACG-UUACUAGAAUUGUUGCACU--UUCCGCG--GGCCCAGAGAGA----UACUUCCGG--AUAAU ..((((((..((..((....(((((((((--.(-(..(((..((.(((((((....-......))))))).)))))--)).))))--))))).....))----..))..)))--))).. ( -30.10, z-score = -0.25, R) >droEre2.scaffold_4784 1473479 97 + 25762168 ----CUUGCCUGGG-----GGGGCCUGUG--AA-AUCAGUUUGUUGCAAUUUGACG-UGACUGGAAUUGUUGCACU--AUUUGCG--GGCCCAGAGAGA----UACUUCCGG-AUUAAU ----.....((((.-----.(((((((..--((-...(((..((.(((((((.(..-....).))))))).)))))--.))..))--))))).(((...----..)))))))-...... ( -28.80, z-score = -0.90, R) >droSec1.super_2 1523515 96 + 7591821 --UGUUUGUUGGGG-------GGCCUGUG--AA-AUCAGUUUGUUGCAAUUUGACA-UGACUGGAAUUGCUGCACU--UUUCGCG--GGCCCAGAGAGA----UACUUCCGG--AUAAU --...(((((.(((-------((((((((--((-(..(((..((.(((((((.(..-....).))))))).)))))--)))))))--))))).(((...----..))))).)--)))). ( -34.90, z-score = -3.06, R) >droSim1.chr3L 1078909 98 + 22553184 UCCGUUUGUUGGGG-------GGCCUGUG--AA-AUCAGUUUGUUGCAAUUUGACA-UGACUGGAAUUGCUGCACU--UUUCGCG--GGCCCAGAGAGA----UACUUCCGG--AUAAU .....(((((.(((-------((((((((--((-(..(((..((.(((((((.(..-....).))))))).)))))--)))))))--))))).(((...----..))))).)--)))). ( -34.90, z-score = -2.51, R) >droAna3.scaffold_13337 21334077 96 - 23293914 ----CUUAUAUUAUU-----UGGCCCGUG---ACAUUA-UUUGUUACAAUUUGUCG-CCUUUGGAAUUGUUGCACGAG-UUCGCG---GGCCCAGAAAGAU----AAUUCCG-GUUAAU ----((...((((((-----(((((((((---(.(((.-..(((.(((((((....-......))))))).)))..))-))))))---)))).....))))----)))...)-)..... ( -25.50, z-score = -1.70, R) >droYak2.chr3L 1446215 98 + 24197627 ----CUUGUUUGGG-----GGGGCCUGUG--AA-AUCAGUUUGCUGCAAUUUGACG-UGACUGGAAUUGUUGCACU--UUUCGCG--GGCCCAGAGAAA----UACUUUCGGUGUUAAU ----..........-----.(((((((((--((-(..(((..((.(((((((.(..-....).))))))).)))))--)))))))--))))).....((----(((.....)))))... ( -32.30, z-score = -2.19, R) >droPer1.super_23 1025085 100 - 1662726 ----GUUGUUGUGU-------GGCCUGUG--AA-AUUA-UUUGUUACAAUUUGUCGCUCGUUGGAAUUGUUGCACUA-CUUUGCG--GGCCCAGAAAGAAC-UUAGAUUCCAUUUUAAU ----(((.((.((.-------((((((..--((-....-..(((.(((((((..(....)...))))))).)))...-.))..))--))))))..)).)))-................. ( -23.80, z-score = -1.09, R) >dp4.chrXR_group3a 861371 100 - 1468910 ----GUUGUUGUGU-------GGCCUGUG--AA-AUUA-UUUGUUACAAUUUGUCGCUCGUUGGAAUUGUUGCACUA-CUUUGCG--GGCCCAGAAAGAAC-UUAGAUUCCAUUUUAAU ----(((.((.((.-------((((((..--((-....-..(((.(((((((..(....)...))))))).)))...-.))..))--))))))..)).)))-................. ( -23.80, z-score = -1.09, R) >droWil1.scaffold_180698 8972110 98 - 11422946 ----GUUGUUUUUG-----GCGCCCUGUU--AAUAUUGUUUGGCUACAAAUUGUU--UCGUUAUUGUUGCCAGAUU--UUUGAUG--GGGCCAGAAUAAA---GAAUUUACG-UAUAAU ----.....(((((-----((.(((.(((--((....(((((((.((((......--......)))).))))))).--.))))))--)))))))))....---.........-...... ( -27.00, z-score = -2.71, R) >droVir3.scaffold_13049 3632989 104 + 25233164 ----ACCGUUUAUUU-----CGCCCCACA--AACAUCA-UUUGCUGCAAUUUGUCG-UGUUUGGAAUUGCAGCAAGCGCUUUGCG-UGGGCCAGAGAAAAUAUGAUAUUCCG-UUUAAU ----...((((.(((-----(..(((((.--.......-.((((((((((((....-......))))))))))))((.....)))-))))...))))))))...........-...... ( -27.60, z-score = -1.27, R) >droMoj3.scaffold_6654 2148651 99 - 2564135 --------UUUAUUU-----CGCCCUACA--AACAUCU-UUCGCUGCAAUUUGUCG-UGUUUGGAAUUGCUGCAAACGCUUUGCAAUGGGCCAGAGUAAAU--GAUAUUCCG-UUUAAU --------....(((-----.((((....--....(((-..(((.((.....)).)-))...)))(((((.((....))...))))))))).))).(((((--(......))-)))).. ( -21.50, z-score = -0.45, R) >droGri2.scaffold_15110 7781992 103 + 24565398 ------AAUUUAUUU-----CGCCCCGCAGAAACAUAA-UUUGUUGCAAUUUGUCG-UGCUUGGAAUUGCGGCAUUCGCUUUACG-UGGGCCAGAGAAAAC-UUAAAUUCCG-UUUAAU ------......(((-----(..(((((..(((.....-..(((((((((((....-......))))))))))).....)))..)-))))...))))....-..........-...... ( -20.30, z-score = 0.66, R) >consensus ____CUUGUUUGGG_______GGCCUGUG__AA_AUCA_UUUGUUGCAAUUUGUCG_UGAUUGGAAUUGCUGCACU__CUUUGCG__GGCCCAGAGAAAA___UACAUUCCG_UUUAAU .....................((((((((............(((.(((((((...........))))))).))).......))))..))))............................ (-10.73 = -9.84 + -0.89)

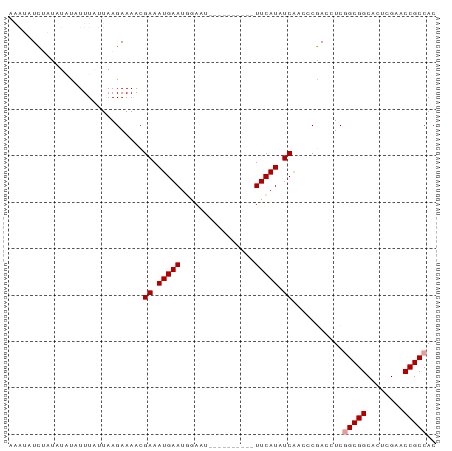
| Location | 1,488,961 – 1,489,056 |
|---|---|
| Length | 95 |
| Sequences | 12 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 68.34 |
| Shannon entropy | 0.62315 |
| G+C content | 0.39211 |
| Mean single sequence MFE | -22.41 |
| Consensus MFE | -8.95 |
| Energy contribution | -8.55 |
| Covariance contribution | -0.40 |
| Combinations/Pair | 1.59 |
| Mean z-score | -1.27 |
| Structure conservation index | 0.40 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.20 |
| SVM RNA-class probability | 0.591944 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
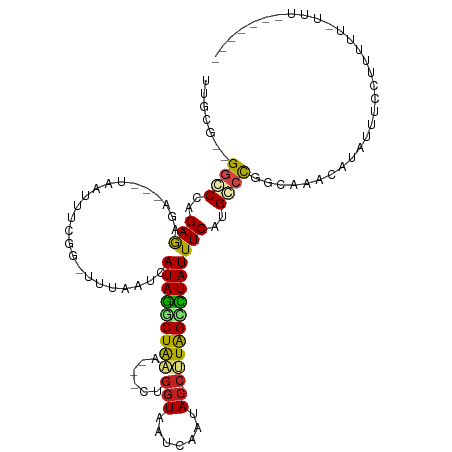
>dm3.chr3L 1488961 95 + 24543557 CCGCG--GGCCCAGAGAGA---UACUUCCGG--AUAAUCAUAGGCUG-----CUGGUUUUCAAUAC-CAUAGUCUAUUUCAUGGCCGGCAAACAAAUUUCCUUUUUGC--------- ..((.--((((..((((((---(........--...))).(((((((-----.((((.......))-)))))))))))))..)))).))...................--------- ( -25.50, z-score = -1.29, R) >droEre2.scaffold_4784 1473544 101 + 25762168 UUGCG--GGCCCAGAGAGA---UACUUCCGG-AUUAAUCAUAGGCUAUAG--CUGGUCAUCAAUAC-CAUAGCUUAUUUCAUGGCCGGCAGACAUAUACCCUUUUUUUUU------- ((((.--((((..((((((---(........-....))).(((((((...--.((((.......))-)))))))))))))..)))).))))...................------- ( -23.70, z-score = -0.74, R) >droSec1.super_2 1523580 89 + 7591821 UCGCG--GGCCCAGAGAGA---UACUUCCGG--AUAAUCAUAGGCUG-----CUGGUUUUCAAUAC-CAUAGCCUAUUUCAUGGCCGGCAAACAAAUUUCCU--------------- ..((.--((((..((((((---(........--...))).(((((((-----.((((.......))-)))))))))))))..)))).)).............--------------- ( -28.20, z-score = -2.34, R) >droSim1.chr3L 1078976 89 + 22553184 UCGCG--GGCCCAGAGAGA---UACUUCCGG--AUAAUCAUAGGCUG-----CUGGUUUUCAAUAC-CAUAGCCUAUUUCAUGGCCGGCAAACAAAUUUCCU--------------- ..((.--((((..((((((---(........--...))).(((((((-----.((((.......))-)))))))))))))..)))).)).............--------------- ( -28.20, z-score = -2.34, R) >droAna3.scaffold_13337 21334142 101 - 23293914 UCGCG--GGCCCAGAAAGA---UAAUUCCGG--UUAAUCAUAGGCUAAAU-CCCGGUAAUCAAUAC-CUUAGUCUAUUUCAUGCCCGGCAAACAUAUUUCUAUUUUUAUU------- ..(((--(((((.(((...---...))).))--......(((((((((..-...((((.....)))-)))))))))).....)))).)).....................------- ( -21.90, z-score = -1.47, R) >droYak2.chr3L 1446280 104 + 24197627 UCGCG--GGCCCAGAGAAA---UACUUUCGGUGUUAAUCAUAGACUAUAUAUCUGGUAAUCAAUAC-CAUAGCCUAUUUCAUGUCCGUUUAACAUAUACCCUUUUUUUGU------- ..(((--(((...((..((---(((.....)))))..))((((.(((......(((((.....)))-))))).)))).....))))))......................------- ( -18.40, z-score = -0.55, R) >droPer1.super_23 1025149 105 - 1662726 UUGCG--GGCCCAGAAAGAACUUAGAUUCCAUUUUAAUCAUAGGCUAAAA--UUGGUAAUCAAUAC-CUUAGUCUAUUUCAUGCAUGGAACACAGUUUUCUUUUUCCUUU------- ....(--((...((((((((((....((((((.......(((((((((..--..((((.....)))-)))))))))).......))))))...)))..))))))))))..------- ( -21.94, z-score = -1.59, R) >dp4.chrXR_group3a 861435 112 - 1468910 UUGCG--GGCCCAGAAAGAACUUAGAUUCCAUUUUAAUCAUAGGCUAAAA--UUGGUAAUCAAUAC-CUUAGUCUAUUUCAUGCAUGGAACACAGUUUUCUUUUUUCUUUUUCCUUU ....(--((...((((((((...(((((((((.......(((((((((..--..((((.....)))-)))))))))).......))))))........)))...))))))))))).. ( -22.54, z-score = -1.63, R) >droWil1.scaffold_180698 8972175 90 - 11422946 UGAUG--GGGCCAGAAUAAA---GAAUUUACGUAUAAUCAUACUCUAA----UGUCUCCAUAAUACGUUUGGAGUAUUUCAUGCCCGUCAAACAUUUUU------------------ (((((--((...........---........((((....))))...((----((.(((((.........))))))))).....))))))).........------------------ ( -18.00, z-score = -1.18, R) >droVir3.scaffold_13049 3633056 106 + 25233164 UUGCG-UGGGCCAGAGAAAAUAUGAUAUUCCGUUUAAUCAUAGACUAAAA--UCGGUGCUCAAUAC-CUUAGCCUAUUUCAUGCCCGGAAUACAUUUUUAUUUUUGUUUU------- ..((.-...))(((((((((.(((.(((((((..((...((((.((((..--..((((.....)))-))))).))))....))..)))))))))).))).))))))....------- ( -19.00, z-score = -0.21, R) >droMoj3.scaffold_6654 2148714 105 - 2564135 UUGCAAUGGGCCAGAGUAAA--UGAUAUUCCGUUUAAUCAUAGGCUAAAU--CGGGUACUCAAUAC-CUUAGCCUAUUUCAUGCCCGGAAUACAGUUUGUUUUUUGAUUU------- ......(((((..((.((((--((......)))))).))((((((((...--.(((((.....)))-)))))))))).....))))).......................------- ( -26.10, z-score = -1.68, R) >droGri2.scaffold_15110 7782059 94 + 24565398 UUACG-UGGGCCAGAGAAAACUUAA-AUUCCGUUUAAUCAUAGACUAAAA--UCGGUACUCAAUAC-CUUAGCCUAUUUCAUGCCCGGAAUACAGAUCU------------------ .....-(((((..((..((((....-.....))))..))((((.((((..--..((((.....)))-))))).)))).....)))))............------------------ ( -15.40, z-score = -0.27, R) >consensus UUGCG__GGCCCAGAGAGA___UAAUUUCGG_UUUAAUCAUAGGCUAAA___CUGGUAAUCAAUAC_CUUAGCCUAUUUCAUGCCCGGCAAACAUAUUUCCUUUUU_UUU_______ .......((((..(((.......................(((((((((.......(....).......))))))))))))..))))............................... ( -8.95 = -8.55 + -0.40)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:55:45 2011