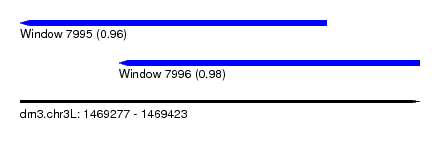
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 1,469,277 – 1,469,423 |
| Length | 146 |
| Max. P | 0.980877 |

| Location | 1,469,277 – 1,469,389 |
|---|---|
| Length | 112 |
| Sequences | 6 |
| Columns | 122 |
| Reading direction | reverse |
| Mean pairwise identity | 69.24 |
| Shannon entropy | 0.53735 |
| G+C content | 0.40183 |
| Mean single sequence MFE | -25.27 |
| Consensus MFE | -17.40 |
| Energy contribution | -16.46 |
| Covariance contribution | -0.94 |
| Combinations/Pair | 1.48 |
| Mean z-score | -1.26 |
| Structure conservation index | 0.69 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.64 |
| SVM RNA-class probability | 0.957387 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 1469277 112 - 24543557 CUUCAAGUCCUUCGAGUUGAAUGCCGAUUUGGAAAUUCAUGGACUUAG---UCCUGCUAAUCCAGUUGGAUUGAAUAUAA-------UACUCGCAUGAGAUGUGGAAUCACAGACAAUAUGA ....(((((((((((((((.....))))))))).......)))))).(---((....((((((....)))))).......-------..(((....))).((((....)))))))....... ( -24.41, z-score = 0.03, R) >droSim1.chr3L 1059735 112 - 22553184 CUGAAAGUCCUUCGAGUGGAAUGCCUAUUUGGAAAUUCAUGGACUUAG---UCCUGCUAAUCCAGGCGGAUUGAAUACCAU---GAAUAGUCGCAUGAAAUGUGGAAUGACAAUAUGA---- ......(((.(((((((((.....))))))))).((((((((..((((---(((.(((......))))))))))...))))---))))..((((((...))))))...))).......---- ( -31.80, z-score = -2.33, R) >droSec1.super_2 1505262 99 - 7591821 CUGAAAGUCCUUCGAGUGGAAUACCUAUUUGGAAAUUCAUGGACUUAGUCCUCCUGCUGAUCCAGCCGGAUUGAAUACCA-------UACUCGCAUGAAAAUAAGA---------------- ((.....((...((((((.....((.....))..(((((.((((...))))(((.((((...)))).))).)))))....-------))))))...)).....)).---------------- ( -22.80, z-score = -1.06, R) >droYak2.chr3L 1427469 102 - 24197627 CUGAAGGUCCUUCGAGUGGAAUGCCUAUUCGGAAAUUCAUGGACUAAG---UCCUGCUAAUCCAGCUGGAUUGAAUACAAUAUGAAAUGUUUG-AUGACAAUGUGA---------------- ......(((.(((((((((.....)))))))))..((((((.....((---(((.(((.....))).)))))........)))))).......-..))).......---------------- ( -23.62, z-score = -0.91, R) >droEre2.scaffold_4784 1454945 95 - 25762168 -------UCCUUCGAGUGGAAUGCCUAUUCGGAAAUUCAUGGACUUAG---UGCUGCUAAUCCAGCUGGAUUGAAUGCAAUAUGAAAUGUUUG-AUGACAAUAUGA---------------- -------...(((((((((.....)))))))))..((((((.......---(((...((((((....))))))...))).)))))).((((..-..))))......---------------- ( -22.40, z-score = -1.15, R) >droAna3.scaffold_13337 21314176 97 + 23293914 CUGAAAGUUUCUCAAGUGAAAUGCUCAUUGGAGAAUUCAUGGACUUAA--GUCCUGCUAAUCCGAUAGGAUUGAAGGCAA-------UGGGGAAAUGGCAAUAUAA---------------- .((...(((((((..(((((...(((....)))..)))))((((....--))))(((((((((....)))))...)))).-------..)))))))..))......---------------- ( -26.60, z-score = -2.11, R) >consensus CUGAAAGUCCUUCGAGUGGAAUGCCUAUUUGGAAAUUCAUGGACUUAG___UCCUGCUAAUCCAGCUGGAUUGAAUACAA_______UACUCGCAUGACAAUAUGA________________ ....(((((((((((((((.....))))))))).......))))))...........((((((....))))))................................................. (-17.40 = -16.46 + -0.94)



| Location | 1,469,313 – 1,469,423 |
|---|---|
| Length | 110 |
| Sequences | 6 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 68.83 |
| Shannon entropy | 0.56753 |
| G+C content | 0.41812 |
| Mean single sequence MFE | -26.20 |
| Consensus MFE | -18.06 |
| Energy contribution | -16.68 |
| Covariance contribution | -1.38 |
| Combinations/Pair | 1.48 |
| Mean z-score | -1.36 |
| Structure conservation index | 0.69 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.06 |
| SVM RNA-class probability | 0.980877 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 1469313 110 - 24543557 ---UAAACGCAAACAGCUGGCAAAUGUUUUUGGUUCGCUUCAAGUCCUUCGAGUUGAAUGCCGAUUUGGAAAUUCAUGGACUUAG---UCCUGCUAAUCCAGUUGGAUUGAAUAUA ---......(((.(((((((.....(((((..(.(((((((((.((....)).))))).).))).)..)))))....((((...)---))).......)))))))..)))...... ( -27.10, z-score = -0.90, R) >droSim1.chr3L 1059771 92 - 22553184 --------------------UAAACG-CUUUGUUUCGCUGAAAGUCCUUCGAGUGGAAUGCCUAUUUGGAAAUUCAUGGACUUAG---UCCUGCUAAUCCAGGCGGAUUGAAUACC --------------------......-.(((..(((((((((..(((...((((((.....)))))))))..))).((((.((((---.....))))))))))))))..))).... ( -21.50, z-score = -0.61, R) >droSec1.super_2 1505282 95 - 7591821 --------------------UAAACG-CUUUGAUUCGCUGAAAGUCCUUCGAGUGGAAUACCUAUUUGGAAAUUCAUGGACUUAGUCCUCCUGCUGAUCCAGCCGGAUUGAAUACC --------------------......-.((..((((((((...(((((((((((((.....))))))))).......))))(((((......)))))..))).)))))..)).... ( -23.31, z-score = -1.43, R) >droYak2.chr3L 1427495 110 - 24197627 ---UAAACGCAAAUAGGUAGUGAAUGUUUUUGGUUCACUGAAGGUCCUUCGAGUGGAAUGCCUAUUCGGAAAUUCAUGGACUAAG---UCCUGCUAAUCCAGCUGGAUUGAAUACA ---............(((((.((.....(((((((((.((((..(((...((((((.....)))))))))..)))))))))))))---)))))))(((((....)))))....... ( -29.40, z-score = -1.39, R) >droEre2.scaffold_4784 1454971 87 - 25762168 --------------------UAAACG----CAAACAGCU--AGUUCCUUCGAGUGGAAUGCCUAUUCGGAAAUUCAUGGACUUAG---UGCUGCUAAUCCAGCUGGAUUGAAUGCA --------------------.....(----((..(((((--(((...(((((((((.....)))))))))......((((.((((---.....))))))))))))).)))..))). ( -25.40, z-score = -1.98, R) >droAna3.scaffold_13337 21314196 114 + 23293914 AUUUUAAACACAGAAUGUCCUGAUGAGUUUAGAUCCCCUGAAAGUUUCUCAAGUGAAAUGCUCAUUGGAGAAUUCAUGGACUUAA--GUCCUGCUAAUCCGAUAGGAUUGAAGGCA ................((((..((((((((...(((..(((..(((((......)))))..)))..)))))))))))))))....--(.(((..((((((....)))))).)))). ( -30.50, z-score = -1.84, R) >consensus ____________________UAAACG_UUUUGAUUCGCUGAAAGUCCUUCGAGUGGAAUGCCUAUUUGGAAAUUCAUGGACUUAG___UCCUGCUAAUCCAGCUGGAUUGAAUACA .........................................(((((((((((((((.....))))))))).......))))))...........((((((....))))))...... (-18.06 = -16.68 + -1.38)


Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:55:41 2011