| Sequence ID | dm3.chr3L |
|---|---|
| Location | 1,423,665 – 1,423,758 |
| Length | 93 |
| Max. P | 0.686319 |

| Location | 1,423,665 – 1,423,758 |
|---|---|
| Length | 93 |
| Sequences | 6 |
| Columns | 104 |
| Reading direction | reverse |
| Mean pairwise identity | 86.85 |
| Shannon entropy | 0.23225 |
| G+C content | 0.37566 |
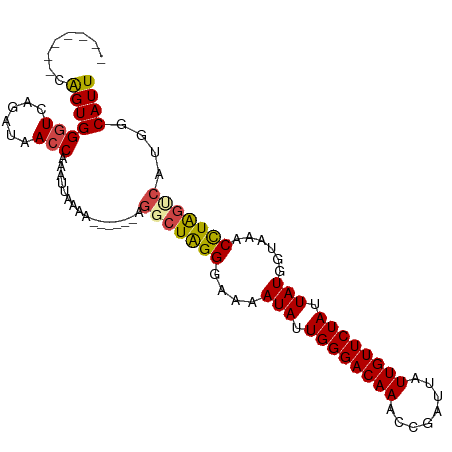
| Mean single sequence MFE | -22.33 |
| Consensus MFE | -18.42 |
| Energy contribution | -18.53 |
| Covariance contribution | 0.11 |
| Combinations/Pair | 1.20 |
| Mean z-score | -1.51 |
| Structure conservation index | 0.82 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.42 |
| SVM RNA-class probability | 0.686319 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
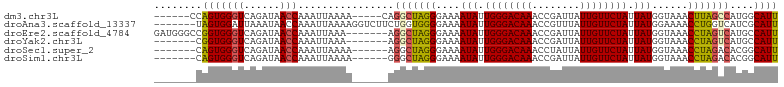
>dm3.chr3L 1423665 93 - 24543557 ------CCAGUGGGUCAGAUAACCAAAUUAAAA-----CAGGCUAGGGAAAAUAUUGGGACAAACCGAUUAUUGUUCUAUUAUGGUAAACUUAGCCAUGGCAUU ------(((...(((......))).........-----..(((((((.(..(((.((((((((........)))))))).)))..)...))))))).))).... ( -23.80, z-score = -1.98, R) >droAna3.scaffold_13337 21272965 97 + 23293914 -------UAGUGGAUUAAAUAACCAAAUUAAAAGGUCUUCUGGUGGGGAAAAUAUUGGGACAAACCGUUUAUUGUUCUAUUAUGGAAAACCUGGUCAUCGCAUU -------..((((....(((......)))...((((.(((((...((((.((((.(((......)))..)))).))))....))))).)))).....))))... ( -16.00, z-score = 0.87, R) >droEre2.scaffold_4784 1412598 97 - 25762168 GAUGGGCCGGUGGGUCAGAUAACCAAAUUAAA-------AGGCUAGGGAAAAUAUUGGGACAAACCGAUUAUUGUUCUAUUAUGGUAAACCUAGUCAUGCCAUU ....(((.(((..........)))........-------.(((((((.(..(((.((((((((........)))))))).)))..)...)))))))..)))... ( -27.00, z-score = -1.72, R) >droYak2.chr3L 1386405 90 - 24197627 -------CGGUGGGUCAGAUAACCAAAUUAAA-------AGGCUAGGGAAAAUAUUGGGACAAACCGAUUAUUGUUCUAUUAUGGUAAACCUAGUCAUGCCAUU -------.(((.(((......)))........-------.(((((((.(..(((.((((((((........)))))))).)))..)...)))))))..)))... ( -25.60, z-score = -2.72, R) >droSec1.super_2 1465586 91 - 7591821 -------CAGUGGGUCAGAUAACCAAAUUAAAA------AGGCUAGGGAAAAUAUUGGGACAAACCUAUUAUUGUUCUAUUAUGGUAAACCUAGACACGGCAUU -------..((((((......))).........------...(((((.(..(((.((((((((........)))))))).)))..)...))))).)))...... ( -20.80, z-score = -1.83, R) >droSim1.chr3L 1019514 91 - 22553184 -------CAGUGGGUCAGAUAACCAAAUUAAAA------GGGCUAGGGAAAAUAUUGGGACAAACCGAUUAUUGUUCUAUUAUGGUAAACCUAGACACGGCAUU -------..((((((......))).........------...(((((.(..(((.((((((((........)))))))).)))..)...))))).)))...... ( -20.80, z-score = -1.71, R) >consensus _______CAGUGGGUCAGAUAACCAAAUUAAAA______AGGCUAGGGAAAAUAUUGGGACAAACCGAUUAUUGUUCUAUUAUGGUAAACCUAGUCAUGGCAUU ........(((((((......)))................(((((((....(((.((((((((........)))))))).)))......)))))))....)))) (-18.42 = -18.53 + 0.11)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:55:35 2011