| Sequence ID | dm3.chr3L |
|---|---|
| Location | 1,413,206 – 1,413,296 |
| Length | 90 |
| Max. P | 0.936995 |

| Location | 1,413,206 – 1,413,296 |
|---|---|
| Length | 90 |
| Sequences | 7 |
| Columns | 96 |
| Reading direction | forward |
| Mean pairwise identity | 74.85 |
| Shannon entropy | 0.49945 |
| G+C content | 0.44913 |
| Mean single sequence MFE | -28.30 |
| Consensus MFE | -8.29 |
| Energy contribution | -9.23 |
| Covariance contribution | 0.94 |
| Combinations/Pair | 1.21 |
| Mean z-score | -3.08 |
| Structure conservation index | 0.29 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.41 |
| SVM RNA-class probability | 0.936995 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
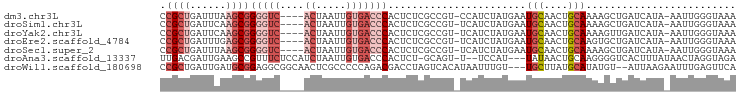
>dm3.chr3L 1413206 90 + 24543557 UUUACCCAAUU-UAUGAUCAGCUUUUGCAGUUGCAUUCAUAGAUGG-ACGGCGAGAGUGGGUCACAAUUAGU----GACCCCGCUUAAAUCAGCGG .....(((.((-(((((.(((((.....)))))...))))))))))-...((....))(((((((.....))----)))))((((......)))). ( -30.10, z-score = -3.51, R) >droSim1.chr3L 1010735 90 + 22553184 UUUACCCAAUU-UAUGAUCAGCUUUUGCAGUUGCAUUCAUAGAUGA-ACGGCGAGAGUGGGUCACAAUUAGU----GACCCCGCUUGAAUCAGCGG ...........-........((((((((.(((.((((....)))))-)).))))))))(((((((.....))----)))))((((......)))). ( -29.90, z-score = -3.45, R) >droYak2.chr3L 1375700 90 + 24197627 UUUACCCAAUU-UAUGAUCAACUUUUGCAGUUGCAUUCAUAGAUGA-ACGGCGAGAGUGGGUCACAAUUAGU----GACCCCGCUUGAAUCAGCGG ...........-........((((((((.(((.((((....)))))-)).))))))))(((((((.....))----)))))((((......)))). ( -29.60, z-score = -3.66, R) >droEre2.scaffold_4784 1402174 90 + 25762168 UUUACCCAAUU-UAUGAUCAGCACUUGCAGUUGCAUUCAUAGAUGA-ACGGCGAGAGUGGGUCACAAUUAGU----GACCCCGCUCAAAUCAGCGG .(..((..(((-(((((.((((.......))))...))))))))..-..))..)(((((((((((.....))----)).))))))).......... ( -28.00, z-score = -3.02, R) >droSec1.super_2 1455147 90 + 7591821 UUUACCCAAUU-UAUGAUCAGCUUUUGCAGUUGCAUUCAUAGAUGA-ACGGCGAGAGUGGGUCACAAUUAGU----GACCCCGCUUAAAUCAGCGG ...........-........((((((((.(((.((((....)))))-)).))))))))(((((((.....))----)))))((((......)))). ( -29.90, z-score = -3.84, R) >droAna3.scaffold_13337 21261364 89 - 23293914 UCUACCUAGUUAUAAAGUGACCCCUUGCAGUUAUA---AUGGA--A-ACUGC-AGAGUGGGUCACAAUUAGAUGGAGAAACGGCUUCAAUCGUCAA ((((.((((((.....((((((((((((((((...---.....--)-)))))-).)).))))))))))))).)))).....(((.......))).. ( -29.90, z-score = -3.83, R) >droWil1.scaffold_180698 8883808 91 - 11422946 UGAACUCAAAUUCUUAAU--ACAUAUGCAUAAGCA---ACAAAUUAUGUGACUAGGUCGUCUGGGGGCGAGUUGCCGCCUCCGCAUCAAUCAGCGG ..................--......((((((...---.....))))))(((......))).(((((((......)))))))((........)).. ( -20.70, z-score = -0.23, R) >consensus UUUACCCAAUU_UAUGAUCAGCUUUUGCAGUUGCAUUCAUAGAUGA_ACGGCGAGAGUGGGUCACAAUUAGU____GACCCCGCUUAAAUCAGCGG ...((((..................(((....)))...............((....))))))..................(((((......))))) ( -8.29 = -9.23 + 0.94)

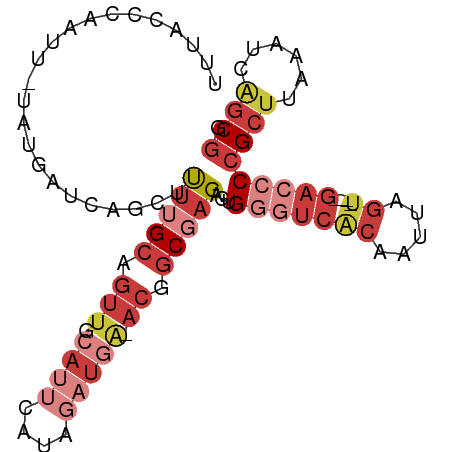
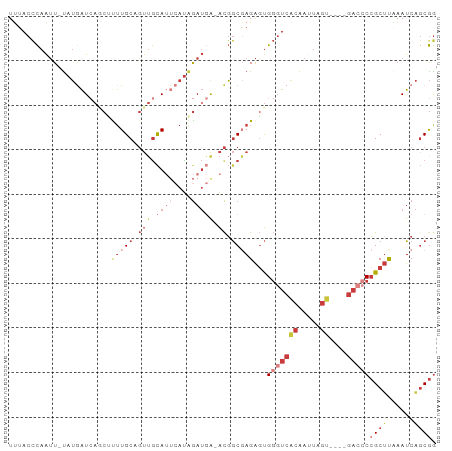
| Location | 1,413,206 – 1,413,296 |
|---|---|
| Length | 90 |
| Sequences | 7 |
| Columns | 96 |
| Reading direction | reverse |
| Mean pairwise identity | 74.85 |
| Shannon entropy | 0.49945 |
| G+C content | 0.44913 |
| Mean single sequence MFE | -26.62 |
| Consensus MFE | -9.07 |
| Energy contribution | -9.77 |
| Covariance contribution | 0.70 |
| Combinations/Pair | 1.31 |
| Mean z-score | -2.56 |
| Structure conservation index | 0.34 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.00 |
| SVM RNA-class probability | 0.870383 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 1413206 90 - 24543557 CCGCUGAUUUAAGCGGGGUC----ACUAAUUGUGACCCACUCUCGCCGU-CCAUCUAUGAAUGCAACUGCAAAAGCUGAUCAUA-AAUUGGGUAAA .((((......))))(((((----((.....)))))))..........(-(((..(((((...((.((.....)).)).)))))-...)))).... ( -26.80, z-score = -2.71, R) >droSim1.chr3L 1010735 90 - 22553184 CCGCUGAUUCAAGCGGGGUC----ACUAAUUGUGACCCACUCUCGCCGU-UCAUCUAUGAAUGCAACUGCAAAAGCUGAUCAUA-AAUUGGGUAAA .((((......))))(((((----((.....)))))))((((..((.((-(((....)))))))....((....))........-....))))... ( -27.00, z-score = -2.75, R) >droYak2.chr3L 1375700 90 - 24197627 CCGCUGAUUCAAGCGGGGUC----ACUAAUUGUGACCCACUCUCGCCGU-UCAUCUAUGAAUGCAACUGCAAAAGUUGAUCAUA-AAUUGGGUAAA .((((......))))(((((----((.....)))))))((((....(((-(((....))))))(((((.....)))))......-....))))... ( -30.00, z-score = -3.81, R) >droEre2.scaffold_4784 1402174 90 - 25762168 CCGCUGAUUUGAGCGGGGUC----ACUAAUUGUGACCCACUCUCGCCGU-UCAUCUAUGAAUGCAACUGCAAGUGCUGAUCAUA-AAUUGGGUAAA .((((......))))(((((----((.....)))))))((((..((.((-(((....)))))))....((....))........-....))))... ( -26.60, z-score = -1.92, R) >droSec1.super_2 1455147 90 - 7591821 CCGCUGAUUUAAGCGGGGUC----ACUAAUUGUGACCCACUCUCGCCGU-UCAUCUAUGAAUGCAACUGCAAAAGCUGAUCAUA-AAUUGGGUAAA .((((......))))(((((----((.....)))))))((((..((.((-(((....)))))))....((....))........-....))))... ( -27.00, z-score = -2.86, R) >droAna3.scaffold_13337 21261364 89 + 23293914 UUGACGAUUGAAGCCGUUUCUCCAUCUAAUUGUGACCCACUCU-GCAGU-U--UCCAU---UAUAACUGCAAGGGGUCACUUUAUAACUAGGUAGA ..((((........))))(((((........(((((((.((.(-(((((-(--.....---...))))))))))))))))..........)).))) ( -26.47, z-score = -3.08, R) >droWil1.scaffold_180698 8883808 91 + 11422946 CCGCUGAUUGAUGCGGAGGCGGCAACUCGCCCCCAGACGACCUAGUCACAUAAUUUGU---UGCUUAUGCAUAUGU--AUUAAGAAUUUGAGUUCA ((((........)))).((.(((.....))).)).(((......)))(((((......---(((....))))))))--.....((((....)))). ( -22.50, z-score = -0.81, R) >consensus CCGCUGAUUUAAGCGGGGUC____ACUAAUUGUGACCCACUCUCGCCGU_UCAUCUAUGAAUGCAACUGCAAAAGCUGAUCAUA_AAUUGGGUAAA (((((......)))))..................(((((.......(((.......)))..(((....))).................)))))... ( -9.07 = -9.77 + 0.70)
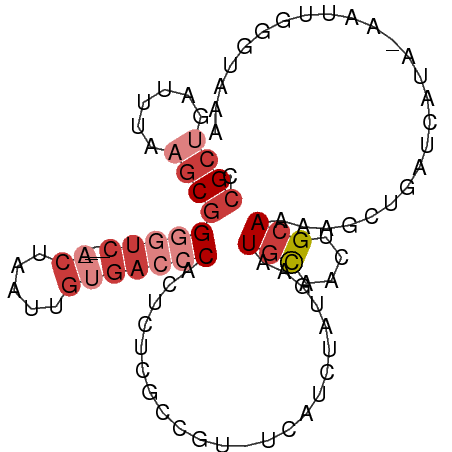
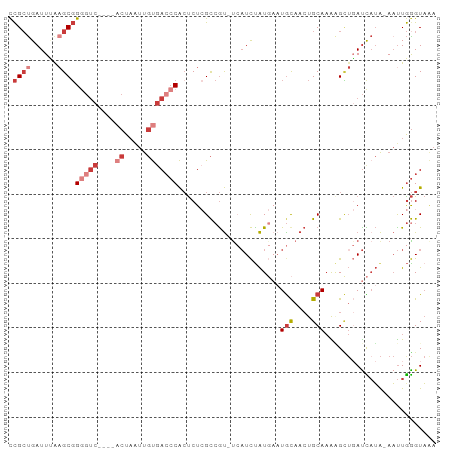
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:55:33 2011