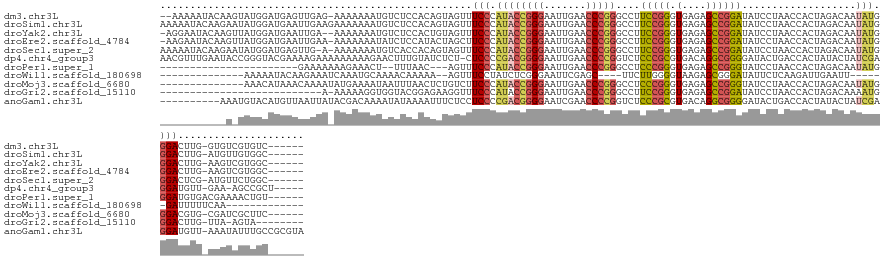
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 1,401,425 – 1,401,559 |
| Length | 134 |
| Max. P | 0.995600 |

| Location | 1,401,425 – 1,401,559 |
|---|---|
| Length | 134 |
| Sequences | 11 |
| Columns | 144 |
| Reading direction | forward |
| Mean pairwise identity | 61.98 |
| Shannon entropy | 0.81516 |
| G+C content | 0.45122 |
| Mean single sequence MFE | -34.31 |
| Consensus MFE | -23.90 |
| Energy contribution | -22.81 |
| Covariance contribution | -1.09 |
| Combinations/Pair | 1.48 |
| Mean z-score | -0.86 |
| Structure conservation index | 0.70 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.82 |
| SVM RNA-class probability | 0.995600 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
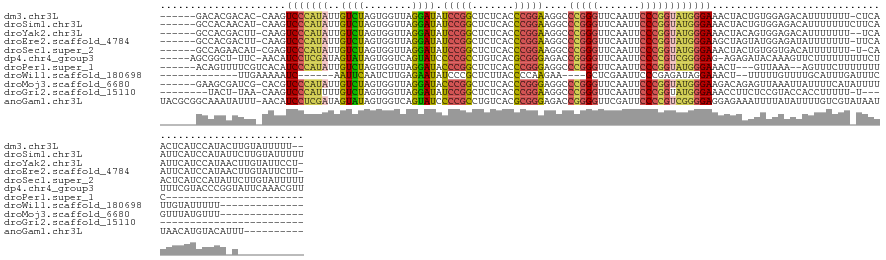
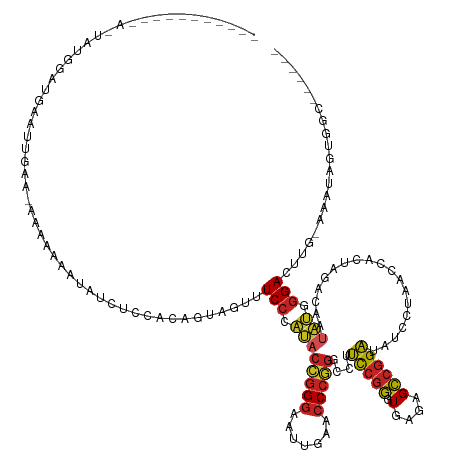
>dm3.chr3L 1401425 134 + 24543557 ------GACACGACAC-CAAGUCCCAUAUUGUCUAGUGGUUAGGAUAUCCGGCUCUCACCCGGAAGGCCCGGGUUCAAUUCCCGGUAUGGGAAACUACUGUGGAGACAUUUUUUU-CUCAACUCAUCCAUACUUGUAUUUUU-- ------..........-(((((.((((........))))...((((..((((.....((((((.....)))))).......))))..(((((((.....(((....)))...)))-))))....))))..))))).......-- ( -36.10, z-score = -0.48, R) >droSim1.chr3L 999092 137 + 22553184 ------GCCACAACAU-CAAGUCCCAUAUUGUCUAGUGGUUAGGAUAUCCGGCUCUCACCCGGAAGGCCCGGGUUCAAUUCCCGGUAUGGGAAACUACUGUGGAGACAUUUUUUUCUUCAAUUCAUCCAUAUUCUUGUAUUUUU ------..........-(((((((((((.(((((........))))).((((.....((((((.....)))))).......)))))))))))........((((((........)))))).............))))....... ( -33.90, z-score = -0.36, R) >droYak2.chr3L 1363724 134 + 24197627 ------GCCACGACUU-CAAGUCCCAUAUUGUCUAGUGGUUAGGAUAUCCGGCUCUCACCCGGAAGGCCCGGGUUCAAUUCCCGGUAUGGGAAACUACAGUGGAGACAUUUUUUU--UCAAUUCAUCCAUAACUUGUAUUCCU- ------(((((.....-(((........)))....)))))..((((..((((.....((((((.....)))))).......))))..((..(((....((((....)))).))).--.))....))))...............- ( -34.60, z-score = -0.22, R) >droEre2.scaffold_4784 1390255 135 + 25762168 ------GCCACGACUU-CAAGUCCCAUAUUGUCUAGUGGUUAGGAUAUCCGGCUCUCACCCGGAAGGCCCGGGUUCAAUUCCCGGUAUGGGAAGCUAGUAUGGAGAUAUUUUUUU-UUCAAUUCAUCCAUAACUUGUAUUCUU- ------(((((.....-(((........)))....))))).(((((((..((((((((.((....)).(((((.......)))))..)))).))))..((((((...(((.....-...)))...))))))....))))))).- ( -36.90, z-score = -0.90, R) >droSec1.super_2 1443730 135 + 7591821 ------GCCAGAACAU-CGAGUCCCAUAUUGUCUAGUGGUUAGGAUAUCCGGCUCUCACCCGGAAGGCCCGGGUUCAAUUCCCGGUAUGGGAAACUACUGUGGUGACAUUUUUUU-U-CAACUCAUCCAUAUUCUUGUAUUUUU ------((.((((...-.(((((((((........))))...((....)))))))((((((((.....(((((.......)))))..(((....)))))).))))).........-.-.............)))).))...... ( -35.60, z-score = -0.37, R) >dp4.chr4_group3 4659386 136 - 11692001 -----AGCGGCU-UUC-AACAUCCUCGAUAGUAUAGUGGUCAGUAUCCCCGCCUGUCACGCGGGAGACCGGGGUUCAAUUCCCCGUCGGGGAG-AGAGAUACAAAGUUCUUUUUUUUUCUUUUCGUACCCGGUAUUCAAACGUU -----..(((..-...-....((((((((........((((......(((((.......))))).))))((((.......))))))))))))(-(((((........))))))...............)))............. ( -38.20, z-score = -0.45, R) >droPer1.super_1 3335960 110 - 10282868 ------ACAGUUUUCGUCACAUCCCAUAUUGUCUAGUGGUUAGGAUACCCGGCUCUCACCCGGGAGGCCCGGGUUCAAUUCCCGGUAUGGGAAACU---GUUAAA--AGUUUCUUUUUUUC----------------------- ------((((((.........((((((((((......((....((.(((((((((((....)))))..))))))))....))))))))))))))))---))....--..............----------------------- ( -33.70, z-score = -1.64, R) >droWil1.scaffold_180698 8871276 105 - 11422946 -------------UUGAAAAAUC------AAUUCAAUCUUGAGAAUAUCCCGCUCUUACCCCAAGAA----GCUCGAAUUCCCGAGAUAGGAAACU--UUUUUGUUUUGCAUUUGAUUUCUUGUAUUUUU-------------- -------------..((.(((((------((....((((((.((((.((..((((((.....))).)----))..)))))).))))))..(((((.--.....)))))....))))))).))........-------------- ( -19.20, z-score = -1.18, R) >droMoj3.scaffold_6680 11862867 123 - 24764193 ------GAAGCGAUCG-CACGUCCCAUAUUGUCUAGUGGUUAGGAUACCCGGCUCUCACCCGGGAGGCCCGGGUUCAAUUCCCGGUAUGGGAAGACAGAGUUAAAUUAUUUUCAUAUUUUGUUUAUGUUU-------------- ------...((....)-)...((((((((((......((....((.(((((((((((....)))))..))))))))....))))))))))))(((((((((..............)))))))))......-------------- ( -37.14, z-score = -1.24, R) >droGri2.scaffold_15110 7687673 106 + 24565398 --------UACU-UAA-CAAGUCCCAUUUUGUCUAGUGGUUAGGAUAUCCGGCUCUCACCCGGAAGGCCCGGGUUCAAUUCCCGGUAUGGGAAACCUUCUCCGUACCACCUUUUU-U--------------------------- --------....-..(-((((......)))))...(((((..(((...((((.....((((((.....)))))).......))))...((....))...)))..)))))......-.--------------------------- ( -31.50, z-score = -1.09, R) >anoGam1.chr3L 6747034 133 + 41284009 UACGCGGCAAAUAUUU-AACAUCCUCGAUAGUAUAGUGGUCAGUAUCCCCGCCUGUCACGCGGGAGACCGGGGUUCGAUUCCCCGUCGGGGAGGAGAAAUUUUAUAUUUUGUCGUAUAAUUAACAUGUACAUUU---------- ...((((((((.((((-....((((((((........((((......(((((.......))))).))))((((.......))))))))))))....)))).......))))))))...................---------- ( -40.60, z-score = -1.51, R) >consensus ______GCCACGAUUU_CAAGUCCCAUAUUGUCUAGUGGUUAGGAUAUCCGGCUCUCACCCGGAAGGCCCGGGUUCAAUUCCCGGUAUGGGAAACUACUGUGGAGACAUUUUUUU_UUCAAUUCAUCCAUA_U___________ .....................(((((((..((((........)))).(((((.......)))))....(((((.......)))))))))))).................................................... (-23.90 = -22.81 + -1.09)
| Location | 1,401,425 – 1,401,559 |
|---|---|
| Length | 134 |
| Sequences | 11 |
| Columns | 144 |
| Reading direction | reverse |
| Mean pairwise identity | 61.98 |
| Shannon entropy | 0.81516 |
| G+C content | 0.45122 |
| Mean single sequence MFE | -34.02 |
| Consensus MFE | -18.18 |
| Energy contribution | -17.55 |
| Covariance contribution | -0.63 |
| Combinations/Pair | 1.35 |
| Mean z-score | -0.81 |
| Structure conservation index | 0.53 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.93 |
| SVM RNA-class probability | 0.975607 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
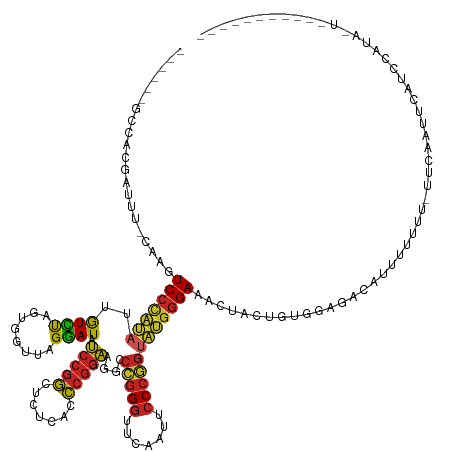
>dm3.chr3L 1401425 134 - 24543557 --AAAAAUACAAGUAUGGAUGAGUUGAG-AAAAAAAUGUCUCCACAGUAGUUUCCCAUACCGGGAAUUGAACCCGGGCCUUCCGGGUGAGAGCCGGAUAUCCUAACCACUAGACAAUAUGGGACUUG-GUGUCGUGUC------ --.......(((((........((.(((-(........)))).))........((((((((((...((..((((((.....))))))..)).)))).....(((.....)))....)))))))))))-..........------ ( -36.40, z-score = -0.27, R) >droSim1.chr3L 999092 137 - 22553184 AAAAAUACAAGAAUAUGGAUGAAUUGAAGAAAAAAAUGUCUCCACAGUAGUUUCCCAUACCGGGAAUUGAACCCGGGCCUUCCGGGUGAGAGCCGGAUAUCCUAACCACUAGACAAUAUGGGACUUG-AUGUUGUGGC------ ...((((((((....((((.(((((.........))).))))))........(((((((((((...((..((((((.....))))))..)).)))).....(((.....)))....)))))))))))-.)))).....------ ( -35.30, z-score = -0.65, R) >droYak2.chr3L 1363724 134 - 24197627 -AGGAAUACAAGUUAUGGAUGAAUUGA--AAAAAAAUGUCUCCACUGUAGUUUCCCAUACCGGGAAUUGAACCCGGGCCUUCCGGGUGAGAGCCGGAUAUCCUAACCACUAGACAAUAUGGGACUUG-AAGUCGUGGC------ -..((...((((...((((.(((((..--.....))).))))))........(((((((((((...((..((((((.....))))))..)).)))).....(((.....)))....)))))))))))-...)).....------ ( -35.60, z-score = -0.09, R) >droEre2.scaffold_4784 1390255 135 - 25762168 -AAGAAUACAAGUUAUGGAUGAAUUGAA-AAAAAAAUAUCUCCAUACUAGCUUCCCAUACCGGGAAUUGAACCCGGGCCUUCCGGGUGAGAGCCGGAUAUCCUAACCACUAGACAAUAUGGGACUUG-AAGUCGUGGC------ -..((...((((.((((((.(((((...-.....))).))))))))......(((((((((((...((..((((((.....))))))..)).)))).....(((.....)))....)))))))))))-...)).....------ ( -38.40, z-score = -1.75, R) >droSec1.super_2 1443730 135 - 7591821 AAAAAUACAAGAAUAUGGAUGAGUUG-A-AAAAAAAUGUCACCACAGUAGUUUCCCAUACCGGGAAUUGAACCCGGGCCUUCCGGGUGAGAGCCGGAUAUCCUAACCACUAGACAAUAUGGGACUCG-AUGUUCUGGC------ .........(((((((((.((((((.-.-.....))).)))))......(..(((((((((((...((..((((((.....))))))..)).)))).....(((.....)))....)))))))..).-)))))))...------ ( -36.90, z-score = -1.01, R) >dp4.chr4_group3 4659386 136 + 11692001 AACGUUUGAAUACCGGGUACGAAAAGAAAAAAAAAGAACUUUGUAUCUCU-CUCCCCGACGGGGAAUUGAACCCCGGUCUCCCGCGUGACAGGCGGGGAUACUGACCACUAUACUAUCGAGGAUGUU-GAA-AGCCGCU----- ...((((.((((..(((((((((................)))))))))..-.(((.(((.((((.......))))((((((((((.......)))))).....)))).........))).)))))))-..)-)))....----- ( -35.49, z-score = 0.22, R) >droPer1.super_1 3335960 110 + 10282868 -----------------------GAAAAAAAGAAACU--UUUAAC---AGUUUCCCAUACCGGGAAUUGAACCCGGGCCUCCCGGGUGAGAGCCGGGUAUCCUAACCACUAGACAAUAUGGGAUGUGACGAAAACUGU------ -----------------------..............--....((---(((((((((((((((...((..(((((((...)))))))..)).)))).....(((.....)))....))))))).........))))))------ ( -33.60, z-score = -1.86, R) >droWil1.scaffold_180698 8871276 105 + 11422946 --------------AAAAAUACAAGAAAUCAAAUGCAAAACAAAAA--AGUUUCCUAUCUCGGGAAUUCGAGC----UUCUUGGGGUAAGAGCGGGAUAUUCUCAAGAUUGAAUU------GAUUUUUCAA------------- --------------.........(((((((((...(((........--.((((..(((((((((((.......----))))))))))).))))(((.....)))....)))..))------)))))))...------------- ( -21.90, z-score = -1.10, R) >droMoj3.scaffold_6680 11862867 123 + 24764193 --------------AAACAUAAACAAAAUAUGAAAAUAAUUUAACUCUGUCUUCCCAUACCGGGAAUUGAACCCGGGCCUCCCGGGUGAGAGCCGGGUAUCCUAACCACUAGACAAUAUGGGACGUG-CGAUCGCUUC------ --------------...((((.......))))....................(((((((((((...((..(((((((...)))))))..)).)))).....(((.....)))....)))))))...(-(....))...------ ( -31.80, z-score = -0.77, R) >droGri2.scaffold_15110 7687673 106 - 24565398 ---------------------------A-AAAAAGGUGGUACGGAGAAGGUUUCCCAUACCGGGAAUUGAACCCGGGCCUUCCGGGUGAGAGCCGGAUAUCCUAACCACUAGACAAAAUGGGACUUG-UUA-AGUA-------- ---------------------------.-.....((((((..(((((((((........(((((.......)))))))))))(.(((....))).)...)))..)))))).(((((........)))-)).-....-------- ( -34.10, z-score = -0.95, R) >anoGam1.chr3L 6747034 133 - 41284009 ----------AAAUGUACAUGUUAAUUAUACGACAAAAUAUAAAAUUUCUCCUCCCCGACGGGGAAUCGAACCCCGGUCUCCCGCGUGACAGGCGGGGAUACUGACCACUAUACUAUCGAGGAUGUU-AAAUAUUUGCCGCGUA ----------..................((((...((((((..(((...(((((......((((.......))))((((((((((.......)))))).....))))...........))))).)))-..))))))....)))) ( -34.70, z-score = -0.64, R) >consensus ___________A_UAUGGAUGAAUUGAA_AAAAAAAUAUCUCCACAGUAGUUUCCCAUACCGGGAAUUGAACCCGGGCCUUCCGGGUGAGAGCCGGAUAUCCUAACCACUAGACAAUAUGGGACUUG_AAAUAGUGGC______ ....................................................(((.((((((((.......)))))....(((((.(....))))))...................))).)))..................... (-18.18 = -17.55 + -0.63)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:55:32 2011