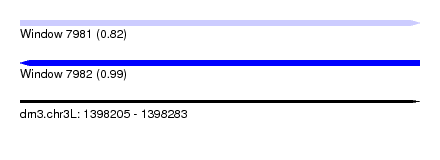
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 1,398,205 – 1,398,283 |
| Length | 78 |
| Max. P | 0.988390 |

| Location | 1,398,205 – 1,398,283 |
|---|---|
| Length | 78 |
| Sequences | 6 |
| Columns | 82 |
| Reading direction | forward |
| Mean pairwise identity | 66.93 |
| Shannon entropy | 0.60849 |
| G+C content | 0.43722 |
| Mean single sequence MFE | -18.48 |
| Consensus MFE | -14.00 |
| Energy contribution | -14.17 |
| Covariance contribution | 0.17 |
| Combinations/Pair | 1.00 |
| Mean z-score | -0.33 |
| Structure conservation index | 0.76 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.80 |
| SVM RNA-class probability | 0.822170 |
| Prediction | RNA |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
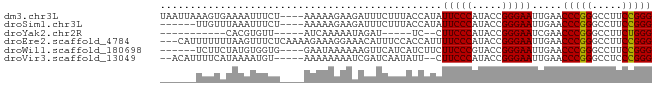
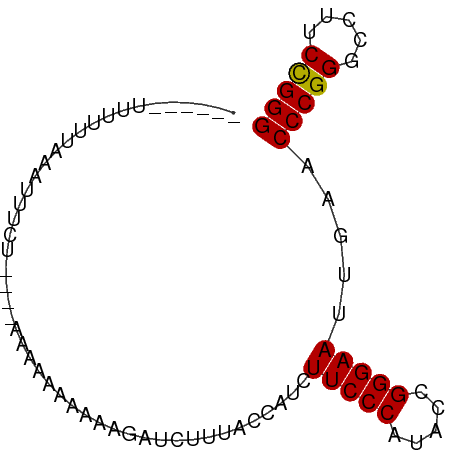
>dm3.chr3L 1398205 78 + 24543557 CCCGGAAGGCCCGGGUUCAAUUCCCGGUAUGGGAAUAUGGUAAAGAAAUCUUCUUUUU----AGAAAUUUUCACUUUAAUUA (((((.....))))).(((((((((.....)))))).)))....((((..((((....----))))..)))).......... ( -21.50, z-score = -1.27, R) >droSim1.chr3L 995910 72 + 22553184 CCCGGAAGGCCCGGGUUCAAUUCCCGGUAUGGGAAUAUGGUAAAGAAAUCUUCUUUUU----AGAAAUUUAAACAA------ (((((.....)))))(((.((((((.....)))))).....(((((.....)))))..----.)))..........------ ( -17.70, z-score = -0.31, R) >droYak2.chr2R 13560842 59 + 21139217 CCCAGAAGGCCCGGGUUCGAUUCCCGGUAUGGGAAG--GA-----AUCUAUUUUUGAU-----AACACGUG----------- ..((((((....((((((..(((((.....))))).--))-----)))).))))))..-----........----------- ( -15.40, z-score = -0.01, R) >droEre2.scaffold_4784 1387025 79 + 25762168 CCCGGAAGGCCCGGGUUCAAUUCCCGGUAUGGGAAAAUGGUGGAAAUGUUUCCUUUCUUUUGAGAAACUUAAAAAAAUG--- (((((.....)))))((((((((((.....)))))......((((....))))......)))))...............--- ( -19.50, z-score = -0.04, R) >droWil1.scaffold_180698 8868144 72 - 11422946 CCCGGAAGGCCCGGGUUCAAUUCCCGGUACGGGAAGAAGAUGAUGAACUUUUUUAUUC----CACCACAUAGAAGA------ ...(((((..(((((.......)))))..)..(((((((........))))))).)))----).............------ ( -18.40, z-score = -0.21, R) >droVir3.scaffold_13049 14391292 73 + 25233164 CCCGGGAGGCCCGGGUUCAAUUCCCGGUAUGGGAAG--AAUAUUGAUCGAUUUUUUUU-----ACAUUUUAUGAAAAUGU-- ((((((...)))))).(((((((((.....))))..--...)))))............-----((((((.....))))))-- ( -18.40, z-score = -0.13, R) >consensus CCCGGAAGGCCCGGGUUCAAUUCCCGGUAUGGGAAGAUGAUAAAGAACUAUUCUUUUU____AAAAACUUAAAAAA______ (((((.....))))).....(((((.....)))))............................................... (-14.00 = -14.17 + 0.17)

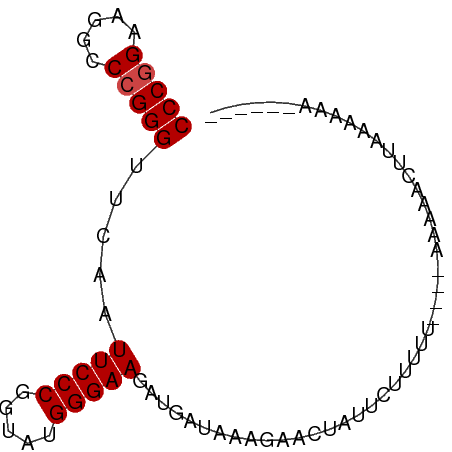
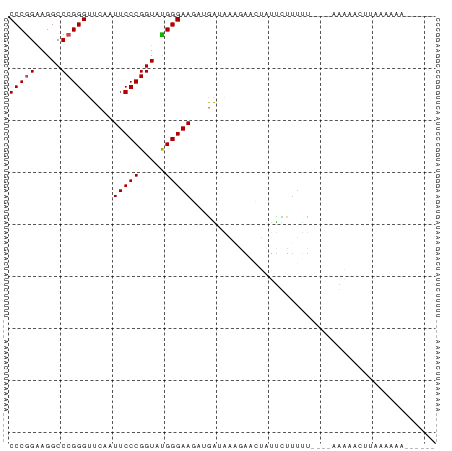
| Location | 1,398,205 – 1,398,283 |
|---|---|
| Length | 78 |
| Sequences | 6 |
| Columns | 82 |
| Reading direction | reverse |
| Mean pairwise identity | 66.93 |
| Shannon entropy | 0.60849 |
| G+C content | 0.43722 |
| Mean single sequence MFE | -18.53 |
| Consensus MFE | -14.62 |
| Energy contribution | -14.48 |
| Covariance contribution | -0.14 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.22 |
| Structure conservation index | 0.79 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.32 |
| SVM RNA-class probability | 0.988390 |
| Prediction | RNA |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
>dm3.chr3L 1398205 78 - 24543557 UAAUUAAAGUGAAAAUUUCU----AAAAAGAAGAUUUCUUUACCAUAUUCCCAUACCGGGAAUUGAACCCGGGCCUUCCGGG ..........((((.(((((----....))))).)))).....((.((((((.....))))))))..(((((.....))))) ( -20.50, z-score = -1.98, R) >droSim1.chr3L 995910 72 - 22553184 ------UUGUUUAAAUUUCU----AAAAAGAAGAUUUCUUUACCAUAUUCCCAUACCGGGAAUUGAACCCGGGCCUUCCGGG ------.........(((((----....)))))..........((.((((((.....))))))))..(((((.....))))) ( -17.40, z-score = -1.05, R) >droYak2.chr2R 13560842 59 - 21139217 -----------CACGUGUU-----AUCAAAAAUAGAU-----UC--CUUCCCAUACCGGGAAUCGAACCCGGGCCUUCUGGG -----------........-----..........(((-----((--((.........)))))))...(((((.....))))) ( -14.00, z-score = -0.18, R) >droEre2.scaffold_4784 1387025 79 - 25762168 ---CAUUUUUUUAAGUUUCUCAAAAGAAAGGAAACAUUUCCACCAUUUUCCCAUACCGGGAAUUGAACCCGGGCCUUCCGGG ---............(((((....)))))((((....))))..((..(((((.....))))).))..(((((.....))))) ( -19.60, z-score = -1.11, R) >droWil1.scaffold_180698 8868144 72 + 11422946 ------UCUUCUAUGUGGUG----GAAUAAAAAAGUUCAUCAUCUUCUUCCCGUACCGGGAAUUGAACCCGGGCCUUCCGGG ------........((((((----((.........))))))))....((((((...)))))).....(((((.....))))) ( -20.20, z-score = -0.81, R) >droVir3.scaffold_13049 14391292 73 - 25233164 --ACAUUUUCAUAAAAUGU-----AAAAAAAAUCGAUCAAUAUU--CUUCCCAUACCGGGAAUUGAACCCGGGCCUCCCGGG --((((((.....))))))-----............(((((...--..((((.....))))))))).((((((...)))))) ( -19.50, z-score = -2.21, R) >consensus ______UUUUUUAAAUUUCU____AAAAAAAAAAGAUCUUUACCAUCUUCCCAUACCGGGAAUUGAACCCGGGCCUUCCGGG ...............................................(((((.....))))).....(((((.....))))) (-14.62 = -14.48 + -0.14)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:55:29 2011