
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 1,397,930 – 1,398,008 |
| Length | 78 |
| Max. P | 0.988953 |

| Location | 1,397,930 – 1,398,008 |
|---|---|
| Length | 78 |
| Sequences | 12 |
| Columns | 97 |
| Reading direction | forward |
| Mean pairwise identity | 82.44 |
| Shannon entropy | 0.36511 |
| G+C content | 0.53524 |
| Mean single sequence MFE | -29.19 |
| Consensus MFE | -28.26 |
| Energy contribution | -27.03 |
| Covariance contribution | -1.23 |
| Combinations/Pair | 1.36 |
| Mean z-score | -1.12 |
| Structure conservation index | 0.97 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.34 |
| SVM RNA-class probability | 0.929230 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
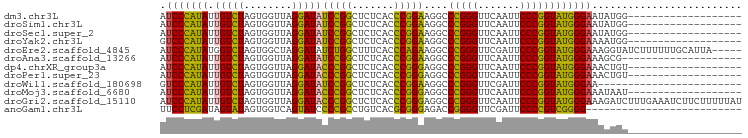
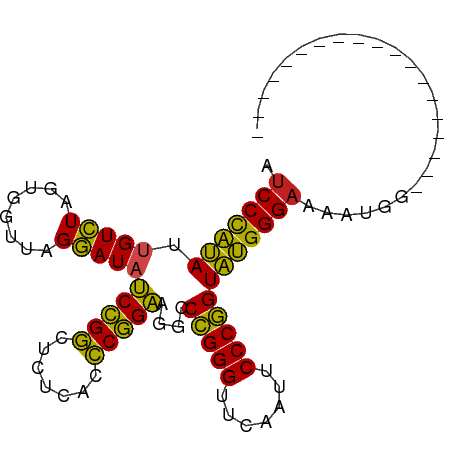
>dm3.chr3L 1397930 78 + 24543557 AUCCCAUAUUGUCUAGUGGUUAGGAUAUCCGGCUCUCACCCGGAAGGCCCGGGUUCAAUUCCCGGUAUGGGAAUAUGG------------------- .(((((((.(((((........))))).((((.....((((((.....)))))).......)))))))))))......------------------- ( -27.60, z-score = -0.83, R) >droSim1.chr3L 995872 78 + 22553184 AUCCCAUAUUGUCUAGUGGUUAGGAUAUCCGGCUCUCACCCGGAAGGCCCGGGUUCAAUUCCCGGUAUGGGAAUAUGG------------------- .(((((((.(((((........))))).((((.....((((((.....)))))).......)))))))))))......------------------- ( -27.60, z-score = -0.83, R) >droSec1.super_2 1440513 78 + 7591821 AUCCCAUAUUGUCUAGUGGUUAGGAUAUCCGGCUCUCACCCGGAAGGCCCGGGUUCAAUUCCCGGUAUGGGAAUAUGG------------------- .(((((((.(((((........))))).((((.....((((((.....)))))).......)))))))))))......------------------- ( -27.60, z-score = -0.83, R) >droYak2.chr3L 1360468 78 + 24197627 GUCCCAUAUUGUCUAGUGGUUAGGAUAUCCGGCUCUCACCCGGAAGGCCCGGGUUCAAUUCCCGGUAUGGGAAAAUGG------------------- .(((((((.(((((........))))).((((.....((((((.....)))))).......)))))))))))......------------------- ( -27.50, z-score = -0.97, R) >droEre2.scaffold_4845 12779967 92 - 22589142 AUCCCAUAUGGUCUAGUGGCUAGGAUAUCUGGCUUUCACCCAGAAGGCCCGGGUUCGAUUCCCGGUAUGGGAAAGGUAUCUUUUUUGCAUUA----- .((((((((((((....)))).........(((((((.....)))))))((((.......))))))))))))...(((.......)))....----- ( -30.00, z-score = -0.93, R) >droAna3.scaffold_13266 10211091 77 - 19884421 AUCCCAUAUUGUCUAGUGGUUAGGAUAUCCGGCUCUCACCCGGAAGGCCCGGGUUCAAUUCCCGGUAUGGGAAAGCG-------------------- .(((((((.(((((........))))).((((.....((((((.....)))))).......))))))))))).....-------------------- ( -27.60, z-score = -1.24, R) >dp4.chrXR_group3a 762406 78 - 1468910 AUCCCAUAUUGUCUAGUGGUUAGGAUACCCGGCUCUCACCCGGGAGGCCCGGGUUCAAUUCCCGGUAUGGGAAACUGU------------------- .((((((((((......((....((.(((((((((((....)))))..))))))))....))))))))))))......------------------- ( -31.00, z-score = -1.35, R) >droPer1.super_23 924533 78 - 1662726 AUCCCAUAUUGUCUAGUGGUUAGGAUACCCGGCUCUCACCCGGGAGGCCCGGGUUCAAUUCCCGGUAUGGGAAACUGU------------------- .((((((((((......((....((.(((((((((((....)))))..))))))))....))))))))))))......------------------- ( -31.00, z-score = -1.35, R) >droWil1.scaffold_180698 8867829 73 - 11422946 GUCCCAUAUUGUCUAGUGGUUAGGAUAUCCGGCUCUCACCCGGAAGGCCCGGGUUCGAUUCCCGGUAUGGGAA------------------------ .(((((((.(((((........))))).((((.((..((((((.....))))))..))...))))))))))).------------------------ ( -28.80, z-score = -1.34, R) >droMoj3.scaffold_6680 11860708 78 - 24764193 AUCCCAUAUUGUCUAGUGGUUAGGAUACCCGGCUCUCACCCGGGAGGCCCGGGUUCAAUUCCCGGUAUGGGAAAUAAU------------------- .((((((((((......((....((.(((((((((((....)))))..))))))))....))))))))))))......------------------- ( -31.00, z-score = -1.61, R) >droGri2.scaffold_15110 8735139 97 - 24565398 AUCCCAUAUUGUCUAGUGGUUAGGAUACCCGGCUCUCACCCGGGAGGCCCGGGUUCAAUUCCCGGUAUGGGAAAGAUCUUUGAAAUCUUCUUUUUAU .((((((((((......((....((.(((((((((((....)))))..))))))))....))))))))))))(((((.......)))))........ ( -31.70, z-score = -0.98, R) >anoGam1.chr3L 6741597 71 + 41284009 UUCCUCGAUAGUAUAGUGGUCAGUAUCCCCGCCUGUCACGCGGGAGACCGGGGUUCGAUUCCCCGUCGGGG-------------------------- ..(((((((........((((......(((((.......))))).))))((((.......)))))))))))-------------------------- ( -28.90, z-score = -1.25, R) >consensus AUCCCAUAUUGUCUAGUGGUUAGGAUAUCCGGCUCUCACCCGGAAGGCCCGGGUUCAAUUCCCGGUAUGGGAAAAUGG___________________ .(((((((.(((((........)))))(((((.......)))))....(((((.......))))))))))))......................... (-28.26 = -27.03 + -1.23)
| Location | 1,397,930 – 1,398,008 |
|---|---|
| Length | 78 |
| Sequences | 12 |
| Columns | 97 |
| Reading direction | reverse |
| Mean pairwise identity | 82.44 |
| Shannon entropy | 0.36511 |
| G+C content | 0.53524 |
| Mean single sequence MFE | -28.97 |
| Consensus MFE | -27.65 |
| Energy contribution | -26.57 |
| Covariance contribution | -1.08 |
| Combinations/Pair | 1.33 |
| Mean z-score | -1.94 |
| Structure conservation index | 0.95 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.34 |
| SVM RNA-class probability | 0.988953 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
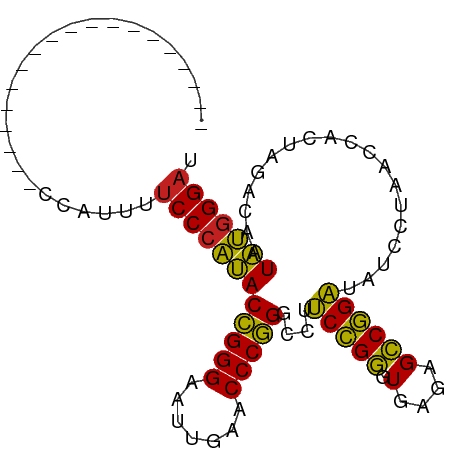
>dm3.chr3L 1397930 78 - 24543557 -------------------CCAUAUUCCCAUACCGGGAAUUGAACCCGGGCCUUCCGGGUGAGAGCCGGAUAUCCUAACCACUAGACAAUAUGGGAU -------------------......(((((((((((...((..((((((.....))))))..)).)))).....(((.....)))....))))))). ( -28.20, z-score = -2.01, R) >droSim1.chr3L 995872 78 - 22553184 -------------------CCAUAUUCCCAUACCGGGAAUUGAACCCGGGCCUUCCGGGUGAGAGCCGGAUAUCCUAACCACUAGACAAUAUGGGAU -------------------......(((((((((((...((..((((((.....))))))..)).)))).....(((.....)))....))))))). ( -28.20, z-score = -2.01, R) >droSec1.super_2 1440513 78 - 7591821 -------------------CCAUAUUCCCAUACCGGGAAUUGAACCCGGGCCUUCCGGGUGAGAGCCGGAUAUCCUAACCACUAGACAAUAUGGGAU -------------------......(((((((((((...((..((((((.....))))))..)).)))).....(((.....)))....))))))). ( -28.20, z-score = -2.01, R) >droYak2.chr3L 1360468 78 - 24197627 -------------------CCAUUUUCCCAUACCGGGAAUUGAACCCGGGCCUUCCGGGUGAGAGCCGGAUAUCCUAACCACUAGACAAUAUGGGAC -------------------......(((((((((((...((..((((((.....))))))..)).)))).....(((.....)))....))))))). ( -28.10, z-score = -2.21, R) >droEre2.scaffold_4845 12779967 92 + 22589142 -----UAAUGCAAAAAAGAUACCUUUCCCAUACCGGGAAUCGAACCCGGGCCUUCUGGGUGAAAGCCAGAUAUCCUAGCCACUAGACCAUAUGGGAU -----....................((((((((((((.......)))))....(((((.(....))))))....((((...))))....))))))). ( -25.70, z-score = -1.26, R) >droAna3.scaffold_13266 10211091 77 + 19884421 --------------------CGCUUUCCCAUACCGGGAAUUGAACCCGGGCCUUCCGGGUGAGAGCCGGAUAUCCUAACCACUAGACAAUAUGGGAU --------------------.....(((((((((((...((..((((((.....))))))..)).)))).....(((.....)))....))))))). ( -28.20, z-score = -2.07, R) >dp4.chrXR_group3a 762406 78 + 1468910 -------------------ACAGUUUCCCAUACCGGGAAUUGAACCCGGGCCUCCCGGGUGAGAGCCGGGUAUCCUAACCACUAGACAAUAUGGGAU -------------------......(((((((((((...((..(((((((...)))))))..)).)))).....(((.....)))....))))))). ( -30.40, z-score = -1.82, R) >droPer1.super_23 924533 78 + 1662726 -------------------ACAGUUUCCCAUACCGGGAAUUGAACCCGGGCCUCCCGGGUGAGAGCCGGGUAUCCUAACCACUAGACAAUAUGGGAU -------------------......(((((((((((...((..(((((((...)))))))..)).)))).....(((.....)))....))))))). ( -30.40, z-score = -1.82, R) >droWil1.scaffold_180698 8867829 73 + 11422946 ------------------------UUCCCAUACCGGGAAUCGAACCCGGGCCUUCCGGGUGAGAGCCGGAUAUCCUAACCACUAGACAAUAUGGGAC ------------------------.(((((((((((...((..((((((.....))))))..)).)))).....(((.....)))....))))))). ( -29.90, z-score = -2.83, R) >droMoj3.scaffold_6680 11860708 78 + 24764193 -------------------AUUAUUUCCCAUACCGGGAAUUGAACCCGGGCCUCCCGGGUGAGAGCCGGGUAUCCUAACCACUAGACAAUAUGGGAU -------------------......(((((((((((...((..(((((((...)))))))..)).)))).....(((.....)))....))))))). ( -30.40, z-score = -1.99, R) >droGri2.scaffold_15110 8735139 97 + 24565398 AUAAAAAGAAGAUUUCAAAGAUCUUUCCCAUACCGGGAAUUGAACCCGGGCCUCCCGGGUGAGAGCCGGGUAUCCUAACCACUAGACAAUAUGGGAU ........((((((.....))))))(((((((((((...((..(((((((...)))))))..)).)))).....(((.....)))....))))))). ( -33.20, z-score = -2.20, R) >anoGam1.chr3L 6741597 71 - 41284009 --------------------------CCCCGACGGGGAAUCGAACCCCGGUCUCCCGCGUGACAGGCGGGGAUACUGACCACUAUACUAUCGAGGAA --------------------------.(((((.((((.......))))((((((((((.......)))))).....)))).........))).)).. ( -26.80, z-score = -1.07, R) >consensus ___________________CCAUUUUCCCAUACCGGGAAUUGAACCCGGGCCUUCCGGGUGAGAGCCGGAUAUCCUAACCACUAGACAAUAUGGGAU .........................((((((((((((.......)))))....(((((.(....))))))...................))))))). (-27.65 = -26.57 + -1.08)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:55:28 2011