| Sequence ID | dm3.chr3L |
|---|---|
| Location | 1,385,863 – 1,385,960 |
| Length | 97 |
| Max. P | 0.989177 |

| Location | 1,385,863 – 1,385,960 |
|---|---|
| Length | 97 |
| Sequences | 9 |
| Columns | 105 |
| Reading direction | reverse |
| Mean pairwise identity | 64.97 |
| Shannon entropy | 0.71614 |
| G+C content | 0.59253 |
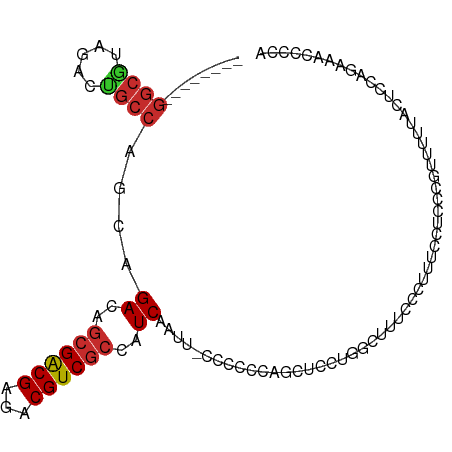
| Mean single sequence MFE | -22.70 |
| Consensus MFE | -12.10 |
| Energy contribution | -12.43 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.25 |
| Mean z-score | -1.35 |
| Structure conservation index | 0.53 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.36 |
| SVM RNA-class probability | 0.989177 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
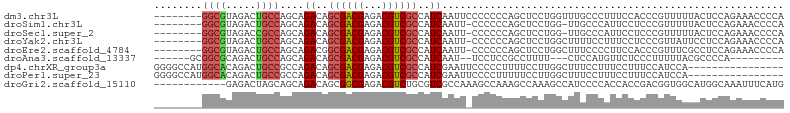
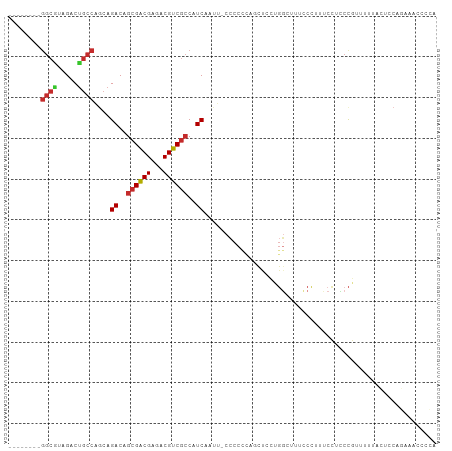
>dm3.chr3L 1385863 97 - 24543557 --------GGCGUAGACUGCCAGCAGACAGCGACGAGACGUCGCCAUCAAUUCCCCCCCAGCUCCUGGUUUGCCCUUUCCACCCGUUUUUACUCCAGAAACCCCA --------((((.....)))).((((((.((((((...))))))..............(((...)))))))))...........((((((.....)))))).... ( -22.10, z-score = -1.89, R) >droSim1.chr3L 983771 95 - 22553184 --------GGCGUAGACUGCCAGCAGACAGCGACGAGACGUCGCCAUCAAUU-CCCCCCAGCUCCUGG-UUGCCCAUUCCUCCCGUUUUUACUCCAGAAACCCCA --------((((.....)))).(((((..((((((...))))))..))....-....((((...))))-.)))...........((((((.....)))))).... ( -21.00, z-score = -1.37, R) >droSec1.super_2 1428494 95 - 7591821 --------GGCGUAGACCGCCAGCAGACAGCGACGAGACGUCGCCAUCAAUU-CCCCCCAGCUCCUGG-UUGCCCAUUCCUCCCGUUUUUACUCCAGAAACCCCA --------((((.....)))).(((((..((((((...))))))..))....-....((((...))))-.)))...........((((((.....)))))).... ( -22.70, z-score = -2.28, R) >droYak2.chr3L 1347917 96 - 24197627 --------GGCGUAGACUGCCAGCAGACAGCGACGAGACGUCGCCAUCAAUU-CCCCCCAGCUCCUGGCUUUUCCUUUCCUCCCGUUAUUCCUCCAGAAACCCCA --------((((.....))))(((.((..((((((...))))))..))....-....((((...))))................))).................. ( -17.70, z-score = -0.27, R) >droEre2.scaffold_4784 1374830 96 - 25762168 --------GGCGUAGACUGCCAGCAGACGGCGACGAGACGUCGCCAUCAAUU-CCCCCCAGCUCCUGGCUUUCCCCUUCCACCCGUUUCGCCUCCAGAAACCCCA --------((((.((((........((.(((((((...))))))).))....-....((((...))))................))))))))............. ( -26.00, z-score = -1.97, R) >droAna3.scaffold_13337 21232606 85 + 23293914 ------GCGGCGCAGACUGCCAGCAGACAGCGACGAGACGUCGCCAUCAAU--UCCUCCGCCUUUU---CUCCAUGUUCUCCCUUUUUACGCCCCA--------- ------..((((.((.(((....)))...((((((...)))))).......--..)).))))....---...........................--------- ( -19.10, z-score = -1.10, R) >dp4.chrXR_group3a 747612 89 + 1468910 GGGGCCAUGGCACAGACUGCCGCCAGACAGCGACGAGACGUCGCCAUCGAAUUCCCCCUUUUCCUUGGCUUUCCUUUCCUUUCCAUCCA---------------- ((((....((((.....))))((((((..((((((...))))))..))(((.........)))..))))...........)))).....---------------- ( -22.70, z-score = -0.57, R) >droPer1.super_23 909728 89 + 1662726 GGGGCCAUGGCACAGACUGCCGCCAGACAGCGACGAGACGUCGCCAUCGAAUUCCCCUUUUUCCUUGGCUUUCCUUUCCUUUCCAUCCA---------------- ((((....((((.....))))((((((..((((((...))))))..))(((.........)))..))))...........)))).....---------------- ( -22.70, z-score = -0.62, R) >droGri2.scaffold_15110 7672482 93 - 24565398 ------------GAGACUAGCAGCAGACAGCGGCGAGACGUCUGCGUCGCCAAAGCCAAAGCCAAAGCCAUCCCCACCACCGACGGUGGCAUGGCAAAUUUCAUG ------------((((...((.((.....))(((..((((....))))......)))...))....(((((..(((((......))))).)))))...))))... ( -30.30, z-score = -2.09, R) >consensus ________GGCGUAGACUGCCAGCAGACAGCGACGAGACGUCGCCAUCAAUU_CCCCCCAGCUCCUGGCUUUCCCUUUCCUCCCGUUUUUACUCCAGAAACCCCA ........((((.....))))....((..((((((...))))))..))......................................................... (-12.10 = -12.43 + 0.33)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:55:26 2011