| Sequence ID | dm3.chr3L |
|---|---|
| Location | 1,380,517 – 1,380,616 |
| Length | 99 |
| Max. P | 0.998630 |

| Location | 1,380,517 – 1,380,616 |
|---|---|
| Length | 99 |
| Sequences | 13 |
| Columns | 110 |
| Reading direction | forward |
| Mean pairwise identity | 68.26 |
| Shannon entropy | 0.71544 |
| G+C content | 0.48079 |
| Mean single sequence MFE | -29.90 |
| Consensus MFE | -22.05 |
| Energy contribution | -21.51 |
| Covariance contribution | -0.54 |
| Combinations/Pair | 1.77 |
| Mean z-score | -0.70 |
| Structure conservation index | 0.74 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.12 |
| SVM RNA-class probability | 0.982932 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
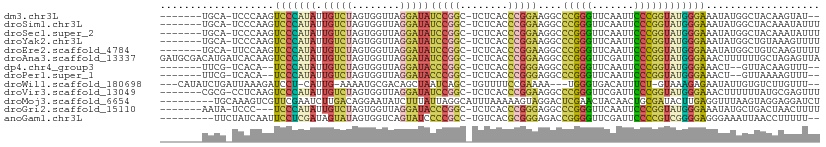
>dm3.chr3L 1380517 99 + 24543557 -------UGCA-UCCCAAGUCCCAUAUUGUCUAGUGGUUAGGAUAUCCGGC-UCUCACCCGGAAGGCCCGGGUUCAAUUCCCGGUAUGGGAAAUAUGGCUACAAGUAU-- -------....-..(((..(((((((.(((((........))))).((((.-....((((((.....)))))).......)))))))))))....)))..........-- ( -29.80, z-score = -0.17, R) >droSim1.chr3L 978436 101 + 22553184 -------UGCA-UCCCAAGUCCCAUAUUGUCUAGUGGUUAGGAUAUCCGGC-UCUCACCCGGAAGGCCCGGGUUCAAUUCCCGGUAUGGGAAAUAUGGCUACAAAUAUUU -------....-..(((..(((((((.(((((........))))).((((.-....((((((.....)))))).......)))))))))))....)))............ ( -29.80, z-score = -0.30, R) >droSec1.super_2 1423170 101 + 7591821 -------UGCA-UCCCAAGUCCCAUAUUGUCUAGUGGUUAGGAUAUCCGGC-UCUCACCCGGAAGGCCCGGGUUCAAUUCCCGGUAUGGGAAAUAUGGCUACAAAUAUUU -------....-..(((..(((((((.(((((........))))).((((.-....((((((.....)))))).......)))))))))))....)))............ ( -29.80, z-score = -0.30, R) >droYak2.chr3L 1341756 101 + 24197627 -------UGCA-UCCCAAGUCCCAUAUUGUCUAGUGGUUAGGAUAUCCGGC-UCUCACCCGGAAGGCCCGGGUUCAAUUCCCGGUAUGGGAAAUAUGGCUGUAAAGUUUU -------((((-.((....(((((((.(((((........))))).((((.-....((((((.....)))))).......))))))))))).....)).))))....... ( -32.60, z-score = -1.03, R) >droEre2.scaffold_4784 1369156 101 + 25762168 -------UGCA-UUCCAAGUCCCAUAUUGUCUAGUGGUUAGGAUAUCCGGC-UCUCACCCGGAAGGCCCGGGUUCAAUUCCCGGUAUGGGAAAUAUGGCUGUCAAGUUUU -------.(((-..(((..(((((((.(((((........))))).((((.-....((((((.....)))))).......)))))))))))....))).)))........ ( -30.50, z-score = -0.51, R) >droAna3.scaffold_13337 21227076 109 - 23293914 GAUGCGACAUGAUCACAAGUCCCAUAUUGUCUAGUGGUUAGGAUAUCCGGC-UCUCACCCGGAAGGCCCGGGUUCGAUUCCCGGUAUGGGAAACUUUUUUGCUAGAGUUA .(((.(((.((....)).))).)))....(((((..(..(((.........-((..((((((.....))))))..))(((((.....))))).)))..)..))))).... ( -32.50, z-score = -0.31, R) >dp4.chr4_group3 6235914 95 - 11692001 -------UUCG-UCACA--UCCCAUAUUGUCUAGUGGUUAGGAUACCCGGC-UCUCACCCGGGAGGCCCGGGUUCAAUUCCCGGUAUGGGAAACU--GUUACAAGUUU-- -------...(-(.(((--((((((((((......((....((.(((((((-((((....)))))..))))))))....))))))))))))...)--)).))......-- ( -33.00, z-score = -1.53, R) >droPer1.super_1 3335966 95 - 10282868 -------UUCG-UCACA--UCCCAUAUUGUCUAGUGGUUAGGAUACCCGGC-UCUCACCCGGGAGGCCCGGGUUCAAUUCCCGGUAUGGGAAACU--GUUAAAAGUUU-- -------....-.....--((((((((((......((....((.(((((((-((((....)))))..))))))))....))))))))))))((((--......)))).-- ( -32.10, z-score = -1.39, R) >droWil1.scaffold_180698 8845246 98 - 11422946 ---CAUAUCUGAUUAAAGAUCCU-CAUUG-AAAAUGCGACAGCUAAUCAGC-UGUUUUCCGAAAA---UGGGUGACAUUUCU-GUAAAGAGAAUAUUGUGUCUUGUUU-- ---...((((......)))).((-((((.-.......(((((((....)))-)))).......))---)))).(((((((((-......))))....)))))......-- ( -19.56, z-score = -0.62, R) >droVir3.scaffold_13049 3495370 101 + 25233164 -------CGCG-CCUCAAGUCCCAUAUUGUCUAGUGGUUAGGAUAUCCGGC-UCUCACCCGGAAGGCCCGGGUUCGAUUCCCGGUAUGGGAAACUUUUUUAUGCGAGUUU -------((((-....((((((((((.(((((........))))).((((.-((..((((((.....))))))..))...)))))))))))..))).....))))..... ( -32.00, z-score = -0.54, R) >droMoj3.scaffold_6654 2034989 101 - 2564135 ---------UGCAAAGUCGUUCGAAUCUUGACAGGAAUAUCUUUAUUAGGCAUUUAAAAAGUAGGACUCGAACUACAACUGCGAUACUUGAGGUUUAAGUAGGAGGAUCU ---------.(((..((.((((((.(((((....((((..((.....))..))))......))))).)))))).))...)))..(((((((...)))))))......... ( -21.70, z-score = -0.74, R) >droGri2.scaffold_15110 8738432 98 - 24565398 -------AAUA-UCCC---UCCCAUAUUGUCUAGUGGUUAGGAUACCCGGC-UCUCACCCGGGAGGCCCGGGUUCAAUUCCCGGUAUGGGAAAUAUGCUGACUAACUUUU -------..((-(((.---..((((........))))...)))))..((((-....(((((((...)))))))....(((((.....)))))....)))).......... ( -31.00, z-score = -0.50, R) >anoGam1.chr3L 4285086 98 + 41284009 ---------UUCUAUCAAUUCCUCGAUAGUAUAGUGGUCAGUAUCCCCGCC-UGUCACGCGGGAGACCGGGGUUCGAUUCCCCGUCGGGGAGGGAAAUUAACCUUUUU-- ---------..(((((........)))))......(((.(((.((((..((-((.(....((....))((((.......))))).))))..)))).))).))).....-- ( -34.40, z-score = -1.11, R) >consensus _______UGCA_UCCCAAGUCCCAUAUUGUCUAGUGGUUAGGAUAUCCGGC_UCUCACCCGGAAGGCCCGGGUUCAAUUCCCGGUAUGGGAAAUAUGGUUACAAGUUUUU ...................(((((((.(((((........)))))(((((........)))))....(((((.......))))))))))))................... (-22.05 = -21.51 + -0.54)
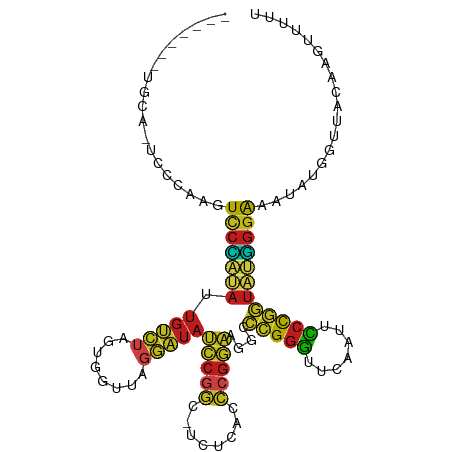
| Location | 1,380,517 – 1,380,616 |
|---|---|
| Length | 99 |
| Sequences | 13 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 68.26 |
| Shannon entropy | 0.71544 |
| G+C content | 0.48079 |
| Mean single sequence MFE | -30.64 |
| Consensus MFE | -21.34 |
| Energy contribution | -21.07 |
| Covariance contribution | -0.27 |
| Combinations/Pair | 1.76 |
| Mean z-score | -1.59 |
| Structure conservation index | 0.70 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.43 |
| SVM RNA-class probability | 0.998630 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 1380517 99 - 24543557 --AUACUUGUAGCCAUAUUUCCCAUACCGGGAAUUGAACCCGGGCCUUCCGGGUGAGA-GCCGGAUAUCCUAACCACUAGACAAUAUGGGACUUGGGA-UGCA------- --.....((((.((.((..(((((((((((...((..((((((.....))))))..))-.)))).....(((.....)))....)))))))..)))).-))))------- ( -33.30, z-score = -1.58, R) >droSim1.chr3L 978436 101 - 22553184 AAAUAUUUGUAGCCAUAUUUCCCAUACCGGGAAUUGAACCCGGGCCUUCCGGGUGAGA-GCCGGAUAUCCUAACCACUAGACAAUAUGGGACUUGGGA-UGCA------- .......((((.((.((..(((((((((((...((..((((((.....))))))..))-.)))).....(((.....)))....)))))))..)))).-))))------- ( -33.30, z-score = -1.57, R) >droSec1.super_2 1423170 101 - 7591821 AAAUAUUUGUAGCCAUAUUUCCCAUACCGGGAAUUGAACCCGGGCCUUCCGGGUGAGA-GCCGGAUAUCCUAACCACUAGACAAUAUGGGACUUGGGA-UGCA------- .......((((.((.((..(((((((((((...((..((((((.....))))))..))-.)))).....(((.....)))....)))))))..)))).-))))------- ( -33.30, z-score = -1.57, R) >droYak2.chr3L 1341756 101 - 24197627 AAAACUUUACAGCCAUAUUUCCCAUACCGGGAAUUGAACCCGGGCCUUCCGGGUGAGA-GCCGGAUAUCCUAACCACUAGACAAUAUGGGACUUGGGA-UGCA------- ............(((....(((((((((((...((..((((((.....))))))..))-.)))).....(((.....)))....)))))))..)))..-....------- ( -31.10, z-score = -1.34, R) >droEre2.scaffold_4784 1369156 101 - 25762168 AAAACUUGACAGCCAUAUUUCCCAUACCGGGAAUUGAACCCGGGCCUUCCGGGUGAGA-GCCGGAUAUCCUAACCACUAGACAAUAUGGGACUUGGAA-UGCA------- ......((....(((....(((((((((((...((..((((((.....))))))..))-.)))).....(((.....)))....)))))))..)))..-..))------- ( -31.30, z-score = -1.75, R) >droAna3.scaffold_13337 21227076 109 + 23293914 UAACUCUAGCAAAAAAGUUUCCCAUACCGGGAAUCGAACCCGGGCCUUCCGGGUGAGA-GCCGGAUAUCCUAACCACUAGACAAUAUGGGACUUGUGAUCAUGUCGCAUC ...................(((((((((((...((..((((((.....))))))..))-.)))).....(((.....)))....)))))))..((((((...)))))).. ( -33.80, z-score = -1.81, R) >dp4.chr4_group3 6235914 95 + 11692001 --AAACUUGUAAC--AGUUUCCCAUACCGGGAAUUGAACCCGGGCCUCCCGGGUGAGA-GCCGGGUAUCCUAACCACUAGACAAUAUGGGA--UGUGA-CGAA------- --....((((.((--(...(((((((((((...((..(((((((...)))))))..))-.)))).....(((.....)))....)))))))--))).)-))).------- ( -33.80, z-score = -1.97, R) >droPer1.super_1 3335966 95 + 10282868 --AAACUUUUAAC--AGUUUCCCAUACCGGGAAUUGAACCCGGGCCUCCCGGGUGAGA-GCCGGGUAUCCUAACCACUAGACAAUAUGGGA--UGUGA-CGAA------- --...........--.((((((((((((((...((..(((((((...)))))))..))-.)))).....(((.....)))....)))))))--...))-)...------- ( -31.30, z-score = -1.44, R) >droWil1.scaffold_180698 8845246 98 + 11422946 --AAACAAGACACAAUAUUCUCUUUAC-AGAAAUGUCACCCA---UUUUCGGAAAACA-GCUGAUUAGCUGUCGCAUUUU-CAAUG-AGGAUCUUUAAUCAGAUAUG--- --......((......((((((.....-.(((((((...((.---.....))...(((-(((....)))))).)))))))-....)-)))))......)).......--- ( -16.40, z-score = -0.70, R) >droVir3.scaffold_13049 3495370 101 - 25233164 AAACUCGCAUAAAAAAGUUUCCCAUACCGGGAAUCGAACCCGGGCCUUCCGGGUGAGA-GCCGGAUAUCCUAACCACUAGACAAUAUGGGACUUGAGG-CGCG------- .....(((......(((..(((((((((((...((..((((((.....))))))..))-.)))).....(((.....)))....))))))))))...)-))..------- ( -33.30, z-score = -1.88, R) >droMoj3.scaffold_6654 2034989 101 + 2564135 AGAUCCUCCUACUUAAACCUCAAGUAUCGCAGUUGUAGUUCGAGUCCUACUUUUUAAAUGCCUAAUAAAGAUAUUCCUGUCAAGAUUCGAACGACUUUGCA--------- .........(((((.......)))))..((((..((.(((((((((.......(((......)))....((((....))))..))))))))).)).)))).--------- ( -18.40, z-score = -1.67, R) >droGri2.scaffold_15110 8738432 98 + 24565398 AAAAGUUAGUCAGCAUAUUUCCCAUACCGGGAAUUGAACCCGGGCCUCCCGGGUGAGA-GCCGGGUAUCCUAACCACUAGACAAUAUGGGA---GGGA-UAUU------- .........((.((....(((((.....)))))((..(((((((...)))))))..))-)).))(((((((..(((.((.....)))))..---))))-))).------- ( -32.70, z-score = -1.09, R) >anoGam1.chr3L 4285086 98 - 41284009 --AAAAAGGUUAAUUUCCCUCCCCGACGGGGAAUCGAACCCCGGUCUCCCGCGUGACA-GGCGGGGAUACUGACCACUAUACUAUCGAGGAAUUGAUAGAA--------- --.....(((((...(((((.((((.((((..((((.....))))..)))))).....-)).)))))...)))))......(((((((....)))))))..--------- ( -36.30, z-score = -2.24, R) >consensus AAAAACUUGUAACCAUAUUUCCCAUACCGGGAAUUGAACCCGGGCCUUCCGGGUGAGA_GCCGGAUAUCCUAACCACUAGACAAUAUGGGACUUGGGA_CGCA_______ ...................((((((((((((.......)))))....(((((........)))))...................)))))))................... (-21.34 = -21.07 + -0.27)

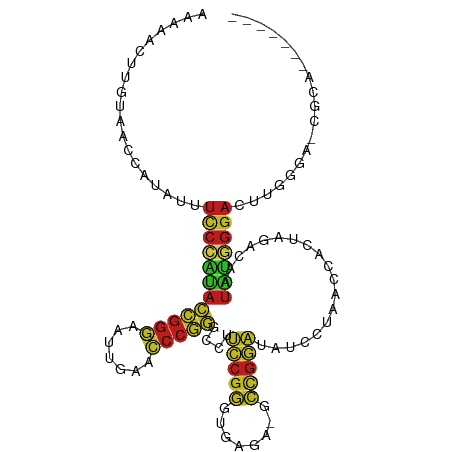
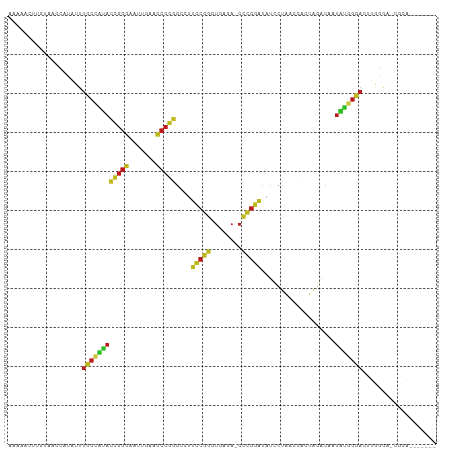
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:55:25 2011