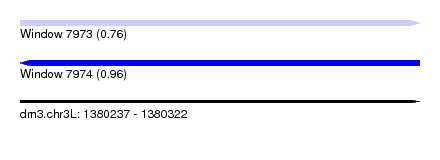
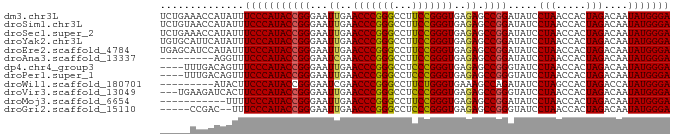
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 1,380,237 – 1,380,322 |
| Length | 85 |
| Max. P | 0.959750 |

| Location | 1,380,237 – 1,380,322 |
|---|---|
| Length | 85 |
| Sequences | 12 |
| Columns | 85 |
| Reading direction | forward |
| Mean pairwise identity | 87.32 |
| Shannon entropy | 0.28730 |
| G+C content | 0.52994 |
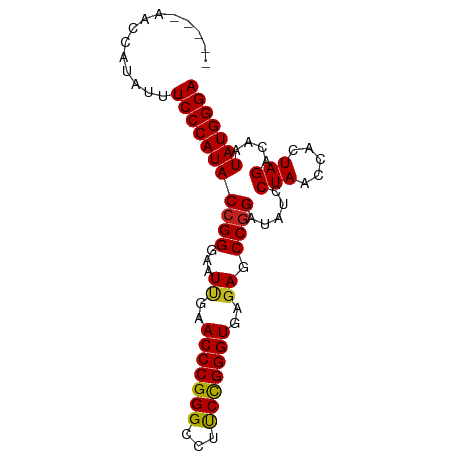
| Mean single sequence MFE | -28.72 |
| Consensus MFE | -27.64 |
| Energy contribution | -27.13 |
| Covariance contribution | -0.51 |
| Combinations/Pair | 1.09 |
| Mean z-score | -0.84 |
| Structure conservation index | 0.96 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.61 |
| SVM RNA-class probability | 0.761869 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
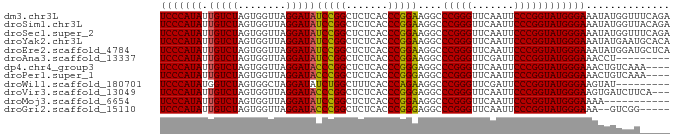
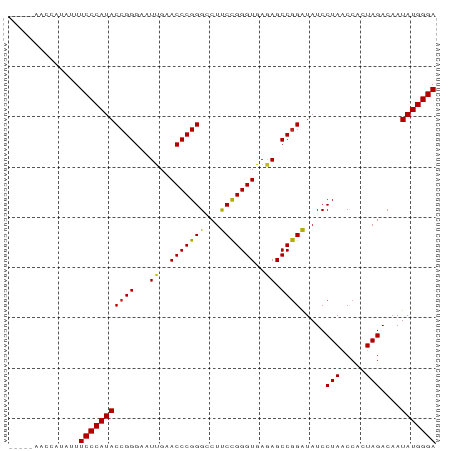
>dm3.chr3L 1380237 85 + 24543557 UCCCAUAUUGUCUAGUGGUUAGGAUAUCCGGCUCUCACCCGGAAGGCCCGGGUUCAAUUCCCGGUAUGGGAAAUAUGGUUUCAGA (((((((.(((((........))))).((((.....((((((.....)))))).......))))))))))).............. ( -27.30, z-score = -0.32, R) >droSim1.chr3L 978187 85 + 22553184 UCCCAUAUUGUCUAGUGGUUAGGAUAUCCGGCUCUCACCCGGAAGGCCCGGGUUCAAUUCCCGGUAUGGGAAAUAUGGUUACAGA (((((((.(((((........))))).((((.....((((((.....)))))).......))))))))))).............. ( -27.30, z-score = -0.29, R) >droSec1.super_2 1422559 85 + 7591821 UCCCAUAUUGUCUAGUGGUUAGGAUAUCCGGCUCUCACCCGGAAGGCCCGGGUUCAAUUCCCGGUAUGGGAAAUAUGGUUUCAGA (((((((.(((((........))))).((((.....((((((.....)))))).......))))))))))).............. ( -27.30, z-score = -0.32, R) >droYak2.chr3L 1341481 85 + 24197627 UCCCAUAUUGUCUAGUGGUUAGGAUAUCCGGCUCUCACCCGGAAGGCCCGGGUUCAAUUCCCGGUAUGGGAAAUAUGAAUGCACA (((((((.(((((........))))).((((.....((((((.....)))))).......))))))))))).............. ( -27.30, z-score = -0.55, R) >droEre2.scaffold_4784 1368886 85 + 25762168 UCCCAUAUUGUCUAGUGGUUAGGAUAUCCGGCUCUCACCCGGAAGGCCCGGGUUCAAUUCCCGGUAUGGGAAAUAUGGAUGCUCA ..((((........))))...(..((((((..(((((.((....)).(((((.......)))))..)))))....))))))..). ( -27.50, z-score = -0.12, R) >droAna3.scaffold_13337 21226795 76 - 23293914 UCCCAUAUUGUCUAGUGGUUAGGAUAUCCGGCUCUCACCCGGAAGGCCCGGGUUCGAUUCCCGGUAUGGGAAACCU--------- (((((((.(((((........))))).((((.((..((((((.....))))))..))...))))))))))).....--------- ( -28.60, z-score = -1.00, R) >dp4.chr4_group3 6235600 81 - 11692001 UCCCAUAUUGUCUAGUGGUUAGGAUACCCGGCUCUCACCCGGGAGGCCCGGGUUCAAUUCCCGGUAUGGGAAACUGUCAAA---- ((((((((((......((....((.(((((((((((....)))))..))))))))....))))))))))))..........---- ( -30.70, z-score = -1.24, R) >droPer1.super_1 3335685 81 - 10282868 UCCCAUAUUGUCUAGUGGUUAGGAUACCCGGCUCUCACCCGGGAGGCCCGGGUUCAAUUCCCGGUAUGGGAAACUGUCAAA---- ((((((((((......((....((.(((((((((((....)))))..))))))))....))))))))))))..........---- ( -30.70, z-score = -1.24, R) >droWil1.scaffold_180701 2122410 76 - 3904529 UCCCAUAUGGUCUAGUGGCUAGGAUAUCUGGCUUUCACCCAGAAGGCCCGGGUUCGAUUCCCGGUAUGGGAAGUAU--------- ((((((((((((....)))).........(((((((.....)))))))((((.......)))))))))))).....--------- ( -29.30, z-score = -1.59, R) >droVir3.scaffold_13049 14388327 82 + 25233164 UCCCAUAUUGUCUAGUGGUUAGGAUACCCGGCUCUCACCCGGGAGGCCCGGGUUCAAUUCCCGGUAUGGGAAGUGAUCUUCA--- ((((((((((......((....((.(((((((((((....)))))..))))))))....))))))))))))...........--- ( -30.70, z-score = -0.95, R) >droMoj3.scaffold_6654 2034662 74 - 2564135 UCCCAUAUUGUCUAGUGGUUAGGAUAUCCGGCUCUCACCCGGAAGGCCCGGGUUCAAUUCCCGGUAUGGGAAAA----------- (((((((.(((((........))))).((((.....((((((.....)))))).......)))))))))))...----------- ( -27.30, z-score = -1.31, R) >droGri2.scaffold_15110 8737798 78 - 24565398 UCCCAUAUUGUCUAGUGGUUAGGAUACCCGGCUCUCACCCGGGAGGCCCGGGUUCAAUUCCCGGUAUGGGAAA--GUCGG----- ((((((((((......((....((.(((((((((((....)))))..))))))))....))))))))))))..--.....----- ( -30.70, z-score = -1.09, R) >consensus UCCCAUAUUGUCUAGUGGUUAGGAUAUCCGGCUCUCACCCGGAAGGCCCGGGUUCAAUUCCCGGUAUGGGAAAUAUGCAU_____ (((((((.(((((........)))))(((((.......)))))....(((((.......)))))))))))).............. (-27.64 = -27.13 + -0.51)
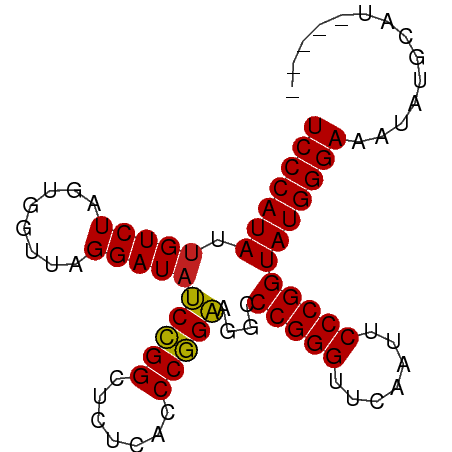
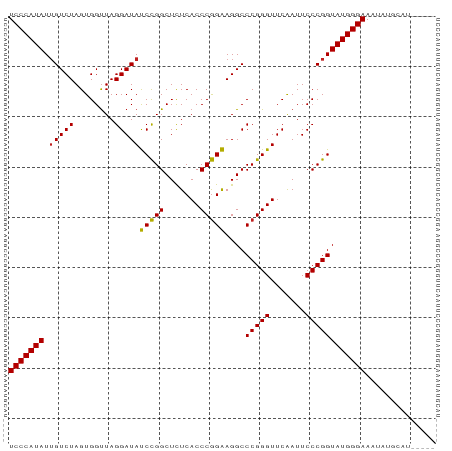
| Location | 1,380,237 – 1,380,322 |
|---|---|
| Length | 85 |
| Sequences | 12 |
| Columns | 85 |
| Reading direction | reverse |
| Mean pairwise identity | 87.32 |
| Shannon entropy | 0.28730 |
| G+C content | 0.52994 |
| Mean single sequence MFE | -28.28 |
| Consensus MFE | -27.55 |
| Energy contribution | -27.35 |
| Covariance contribution | -0.20 |
| Combinations/Pair | 1.13 |
| Mean z-score | -1.49 |
| Structure conservation index | 0.97 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.67 |
| SVM RNA-class probability | 0.959750 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 1380237 85 - 24543557 UCUGAAACCAUAUUUCCCAUACCGGGAAUUGAACCCGGGCCUUCCGGGUGAGAGCCGGAUAUCCUAACCACUAGACAAUAUGGGA ..............(((((((((((...((..((((((.....))))))..)).)))).....(((.....)))....))))))) ( -27.60, z-score = -1.52, R) >droSim1.chr3L 978187 85 - 22553184 UCUGUAACCAUAUUUCCCAUACCGGGAAUUGAACCCGGGCCUUCCGGGUGAGAGCCGGAUAUCCUAACCACUAGACAAUAUGGGA ..............(((((((((((...((..((((((.....))))))..)).)))).....(((.....)))....))))))) ( -27.60, z-score = -1.32, R) >droSec1.super_2 1422559 85 - 7591821 UCUGAAACCAUAUUUCCCAUACCGGGAAUUGAACCCGGGCCUUCCGGGUGAGAGCCGGAUAUCCUAACCACUAGACAAUAUGGGA ..............(((((((((((...((..((((((.....))))))..)).)))).....(((.....)))....))))))) ( -27.60, z-score = -1.52, R) >droYak2.chr3L 1341481 85 - 24197627 UGUGCAUUCAUAUUUCCCAUACCGGGAAUUGAACCCGGGCCUUCCGGGUGAGAGCCGGAUAUCCUAACCACUAGACAAUAUGGGA ..............(((((((((((...((..((((((.....))))))..)).)))).....(((.....)))....))))))) ( -27.60, z-score = -1.24, R) >droEre2.scaffold_4784 1368886 85 - 25762168 UGAGCAUCCAUAUUUCCCAUACCGGGAAUUGAACCCGGGCCUUCCGGGUGAGAGCCGGAUAUCCUAACCACUAGACAAUAUGGGA ..............(((((((((((...((..((((((.....))))))..)).)))).....(((.....)))....))))))) ( -27.60, z-score = -1.27, R) >droAna3.scaffold_13337 21226795 76 + 23293914 ---------AGGUUUCCCAUACCGGGAAUCGAACCCGGGCCUUCCGGGUGAGAGCCGGAUAUCCUAACCACUAGACAAUAUGGGA ---------.....(((((((((((...((..((((((.....))))))..)).)))).....(((.....)))....))))))) ( -29.40, z-score = -1.62, R) >dp4.chr4_group3 6235600 81 + 11692001 ----UUUGACAGUUUCCCAUACCGGGAAUUGAACCCGGGCCUCCCGGGUGAGAGCCGGGUAUCCUAACCACUAGACAAUAUGGGA ----..........(((((((((((...((..(((((((...)))))))..)).)))).....(((.....)))....))))))) ( -29.80, z-score = -1.47, R) >droPer1.super_1 3335685 81 + 10282868 ----UUUGACAGUUUCCCAUACCGGGAAUUGAACCCGGGCCUCCCGGGUGAGAGCCGGGUAUCCUAACCACUAGACAAUAUGGGA ----..........(((((((((((...((..(((((((...)))))))..)).)))).....(((.....)))....))))))) ( -29.80, z-score = -1.47, R) >droWil1.scaffold_180701 2122410 76 + 3904529 ---------AUACUUCCCAUACCGGGAAUCGAACCCGGGCCUUCUGGGUGAAAGCCAGAUAUCCUAGCCACUAGACCAUAUGGGA ---------.....((((((((((((.......)))))....(((((.(....))))))....((((...))))....))))))) ( -25.10, z-score = -1.42, R) >droVir3.scaffold_13049 14388327 82 - 25233164 ---UGAAGAUCACUUCCCAUACCGGGAAUUGAACCCGGGCCUCCCGGGUGAGAGCCGGGUAUCCUAACCACUAGACAAUAUGGGA ---...........(((((((((((...((..(((((((...)))))))..)).)))).....(((.....)))....))))))) ( -29.80, z-score = -1.45, R) >droMoj3.scaffold_6654 2034662 74 + 2564135 -----------UUUUCCCAUACCGGGAAUUGAACCCGGGCCUUCCGGGUGAGAGCCGGAUAUCCUAACCACUAGACAAUAUGGGA -----------...(((((((((((...((..((((((.....))))))..)).)))).....(((.....)))....))))))) ( -27.60, z-score = -2.09, R) >droGri2.scaffold_15110 8737798 78 + 24565398 -----CCGAC--UUUCCCAUACCGGGAAUUGAACCCGGGCCUCCCGGGUGAGAGCCGGGUAUCCUAACCACUAGACAAUAUGGGA -----.....--..(((((((((((...((..(((((((...)))))))..)).)))).....(((.....)))....))))))) ( -29.80, z-score = -1.47, R) >consensus _____AACCAUAUUUCCCAUACCGGGAAUUGAACCCGGGCCUUCCGGGUGAGAGCCGGAUAUCCUAACCACUAGACAAUAUGGGA ..............(((((((((((...((..(((((((...)))))))..)).)))).....(((.....)))....))))))) (-27.55 = -27.35 + -0.20)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:55:23 2011