| Sequence ID | dm3.chr3L |
|---|---|
| Location | 1,379,933 – 1,380,027 |
| Length | 94 |
| Max. P | 0.915278 |

| Location | 1,379,933 – 1,380,027 |
|---|---|
| Length | 94 |
| Sequences | 11 |
| Columns | 96 |
| Reading direction | reverse |
| Mean pairwise identity | 81.22 |
| Shannon entropy | 0.41361 |
| G+C content | 0.49621 |
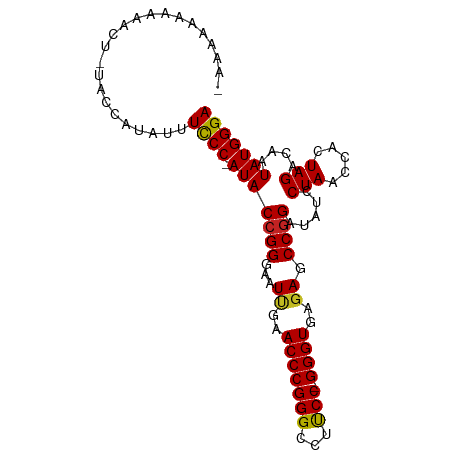
| Mean single sequence MFE | -29.45 |
| Consensus MFE | -20.46 |
| Energy contribution | -19.85 |
| Covariance contribution | -0.62 |
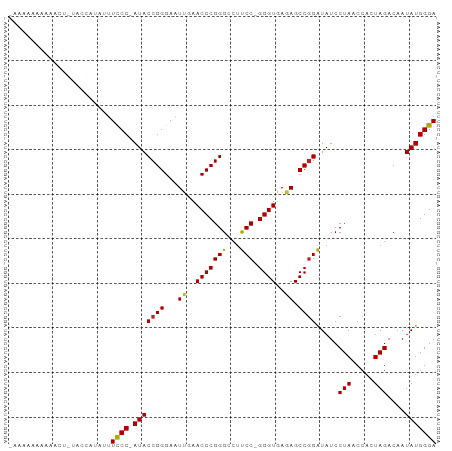
| Combinations/Pair | 1.11 |
| Mean z-score | -1.64 |
| Structure conservation index | 0.69 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.24 |
| SVM RNA-class probability | 0.915278 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
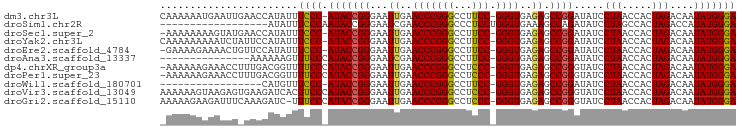
>dm3.chr3L 1379933 94 - 24543557 CAAAAAAUGAAUUGAACCAUAUUUCCC-AUACCGGGAAUUGAACCCGGGCCUUCC-GGGUGAGAGCCGGAUAUCCUAACCACUAGACAAUAUGGGA .......................((((-(((((((...((..((((((.....))-))))..)).)))).....(((.....)))....))))))) ( -27.60, z-score = -1.59, R) >droSim1.chr2R 5287108 78 + 19596830 ------------------AUAUUUCCCAAUACCGGGAAUCGAACCCGGGCCCUCCUGGGUGAAAGCCAGAUAUCCUAGCCACUAGACCAUAUGGGA ------------------.....(((((...(((((.......)))))((((....))))....((.((.....)).))............))))) ( -23.10, z-score = -0.48, R) >droSec1.super_2 1422225 93 - 7591821 -AAAAAAAAGUAUGAACCAUAUUUCCC-AUACCGGGAAUUGAACCCGGGCCUUCC-GGGUGAGAGCCGGAUAUCCUAACCACUAGACAAUAUGGGA -......(((((((...)))))))(((-(((((((...((..((((((.....))-))))..)).)))).....(((.....)))....)))))). ( -28.30, z-score = -1.73, R) >droYak2.chr3L 1341154 94 - 24197627 CAAAAAAAAAUCUAUUCCAUAUUUCCC-AUACCGGGAAUUGAACCCGGGCCUUCC-GGGUGAGAGCCGGAUAUCCUAACCACUAGACAAUAUGGGA .......................((((-(((((((...((..((((((.....))-))))..)).)))).....(((.....)))....))))))) ( -27.60, z-score = -1.56, R) >droEre2.scaffold_4784 1368570 93 - 25762168 -GAAAAGAAAACUGUUCCAUAUUUCCC-AUACCGGGAAUUGAACCCGGGCCUUCC-GGGUGAGAGCCGGAUAUCCUAACCACUAGACAAUAUGGGA -......................((((-(((((((...((..((((((.....))-))))..)).)))).....(((.....)))....))))))) ( -27.60, z-score = -1.19, R) >droAna3.scaffold_13337 21226522 80 + 23293914 ---------------AAAAAAGUUUCCCAUACCGGGAAUCGAACCCGGGCCUUCC-GGGUGAGAGCCGGAUAUCCUAACCACUAGACAAUAUGGGA ---------------.........(((((((((((...((..((((((.....))-))))..)).)))).....(((.....)))....))))))) ( -29.40, z-score = -2.26, R) >dp4.chrXR_group3a 741915 94 + 1468910 -AAAAAAGAAACCUUUGACGGUUUUCCCAUACCGGGAAUUGAACCCGGGCCUCCC-GGGUGAGAGCCGGGUAUCCUAACCACUAGACAAUAUGGGA -......((((((......))))))((((((((((...((..(((((((...)))-))))..)).)))).....(((.....)))....)))))). ( -33.40, z-score = -1.70, R) >droPer1.super_23 903752 94 + 1662726 -AAAAAAGAAACCUUUGACGGUUUUCCCAUACCGGGAAUUGAACCCGGGCCUCCC-GGGUGAGAGCCGGGUAUCCUAACCACUAGACAAUAUGGGA -......((((((......))))))((((((((((...((..(((((((...)))-))))..)).)))).....(((.....)))....)))))). ( -33.40, z-score = -1.70, R) >droWil1.scaffold_180701 2121840 77 + 3904529 -----------------CAUGUUUCCC-AUACCGGGAAUUGAACCCGGGCCUUCC-GGGUGAGAGCCGGAUAUCCUAACCACUAGACAAUAUGGGA -----------------......((((-(((((((...((..((((((.....))-))))..)).)))).....(((.....)))....))))))) ( -27.60, z-score = -1.76, R) >droVir3.scaffold_13049 14388327 95 - 25233164 AAAAAAGUAAGAGUGAAGAUCACUUCCCAUACCGGGAAUUGAACCCGGGCCUCCC-GGGUGAGAGCCGGGUAUCCUAACCACUAGACAAUAUGGGA ..........(((((.....)))))((((((((((...((..(((((((...)))-))))..)).)))).....(((.....)))....)))))). ( -33.00, z-score = -2.04, R) >droGri2.scaffold_15110 8735140 94 + 24565398 AAAAAGAAGAUUUCAAAGAUC-UUUCCCAUACCGGGAAUUGAACCCGGGCCUCCC-GGGUGAGAGCCGGGUAUCCUAACCACUAGACAAUAUGGGA .....(((((((.....))))-)))((((((((((...((..(((((((...)))-))))..)).)))).....(((.....)))....)))))). ( -32.90, z-score = -2.03, R) >consensus _AAAAAAAAAACU_UACCAUAUUUCCC_AUACCGGGAAUUGAACCCGGGCCUUCC_GGGUGAGAGCCGGAUAUCCUAACCACUAGACAAUAUGGGA .......................((((.((((((((.......)))))....(((..(((....))))))...................))))))) (-20.46 = -19.85 + -0.62)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:55:22 2011