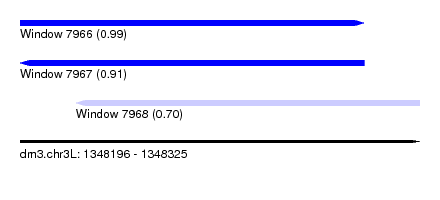
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 1,348,196 – 1,348,325 |
| Length | 129 |
| Max. P | 0.989386 |

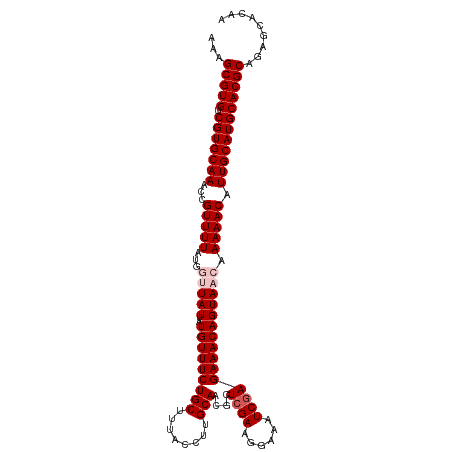
| Location | 1,348,196 – 1,348,307 |
|---|---|
| Length | 111 |
| Sequences | 5 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 95.03 |
| Shannon entropy | 0.08435 |
| G+C content | 0.40606 |
| Mean single sequence MFE | -33.88 |
| Consensus MFE | -30.52 |
| Energy contribution | -31.52 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.93 |
| Structure conservation index | 0.90 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.37 |
| SVM RNA-class probability | 0.989386 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 1348196 111 + 24543557 AAAGCGUGUCGUGCAAACCGUUUUAUGGUUAUUAUGUUUCUGCUUUACCUUGCAACGCUCGAAGGAAAUCGAGAAACAGUAACAAAAACAUUGCAUGCACGCAGAGCACAA ...(((((.(((((((...(((((...(((((..(((((((((........)))....((((......))))))))))))))).))))).))))))))))))......... ( -34.20, z-score = -2.71, R) >droSim1.chr3L 947181 111 + 22553184 AAAGCGUGUCGUGCAAACCGUUUUAUGGUUAUUAUGUUUCUGCUUUACCUUGCAACGCUCGAAGGAAAUCGAGAAACAGUAACAAAAACAUUGCAUGCACGCAGAGCACAA ...(((((.(((((((...(((((...(((((..(((((((((........)))....((((......))))))))))))))).))))).))))))))))))......... ( -34.20, z-score = -2.71, R) >droSec1.super_2 1390735 111 + 7591821 AAAGCGUGUCGUGCAAACCGUUUUAUGGUUAUUAUGUUUCUGCUUUACCUUGCAACGCUCGAAGGAAAUCGAGAAACAGUAACAAAAACAUUGCAUGCACGCAGAGCACAA ...(((((.(((((((...(((((...(((((..(((((((((........)))....((((......))))))))))))))).))))).))))))))))))......... ( -34.20, z-score = -2.71, R) >droEre2.scaffold_4784 1330976 108 + 25762168 AAAGCGUGUCGUGCAAACAGUUUUAU---UAUUAUGUUUCUGCUUUACCUUGCAACGCUCGAGGAAAAUCGAGAAACAGUAACAAAAACAUUGCAUGCACGCAGAGCAUAA ...(((((.(((((((...(((((..---.....((((((((.(((.(((((.......))))).))).).)))))))......))))).))))))))))))......... ( -32.02, z-score = -2.93, R) >droYak2.chr3L 1307229 108 + 24197627 AAAGCGUGUCGUGCAAUCGGUUUUAU---UAUUAUGUUUCUGCUUUACCUUGCAACGCUCGAGGAAAAUCAAGAAACAGUAACAAAAACGUUGCAUGCACGCAGAGCAUAA ...(((((.(((((((.((.((((.(---(((..(((((((..(((.(((((.......))))).)))...))))))))))).)))).))))))))))))))......... ( -34.80, z-score = -3.57, R) >consensus AAAGCGUGUCGUGCAAACCGUUUUAUGGUUAUUAUGUUUCUGCUUUACCUUGCAACGCUCGAAGGAAAUCGAGAAACAGUAACAAAAACAUUGCAUGCACGCAGAGCACAA ...(((((.(((((((...(((((...(((((..(((((((((........)))....((((......))))))))))))))).))))).))))))))))))......... (-30.52 = -31.52 + 1.00)

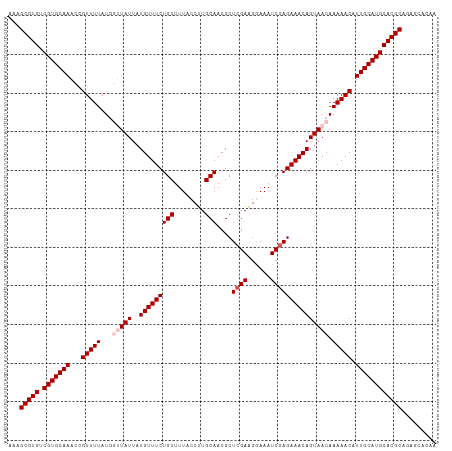
| Location | 1,348,196 – 1,348,307 |
|---|---|
| Length | 111 |
| Sequences | 5 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 95.03 |
| Shannon entropy | 0.08435 |
| G+C content | 0.40606 |
| Mean single sequence MFE | -31.34 |
| Consensus MFE | -28.32 |
| Energy contribution | -28.16 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.05 |
| Structure conservation index | 0.90 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.19 |
| SVM RNA-class probability | 0.906916 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 1348196 111 - 24543557 UUGUGCUCUGCGUGCAUGCAAUGUUUUUGUUACUGUUUCUCGAUUUCCUUCGAGCGUUGCAAGGUAAAGCAGAAACAUAAUAACCAUAAAACGGUUUGCACGACACGCUUU .........((((((.((((((((((((((((((((..(((((......)))))....))..))))..))))))))))...((((.......)))))))).).)))))... ( -32.30, z-score = -2.01, R) >droSim1.chr3L 947181 111 - 22553184 UUGUGCUCUGCGUGCAUGCAAUGUUUUUGUUACUGUUUCUCGAUUUCCUUCGAGCGUUGCAAGGUAAAGCAGAAACAUAAUAACCAUAAAACGGUUUGCACGACACGCUUU .........((((((.((((((((((((((((((((..(((((......)))))....))..))))..))))))))))...((((.......)))))))).).)))))... ( -32.30, z-score = -2.01, R) >droSec1.super_2 1390735 111 - 7591821 UUGUGCUCUGCGUGCAUGCAAUGUUUUUGUUACUGUUUCUCGAUUUCCUUCGAGCGUUGCAAGGUAAAGCAGAAACAUAAUAACCAUAAAACGGUUUGCACGACACGCUUU .........((((((.((((((((((((((((((((..(((((......)))))....))..))))..))))))))))...((((.......)))))))).).)))))... ( -32.30, z-score = -2.01, R) >droEre2.scaffold_4784 1330976 108 - 25762168 UUAUGCUCUGCGUGCAUGCAAUGUUUUUGUUACUGUUUCUCGAUUUUCCUCGAGCGUUGCAAGGUAAAGCAGAAACAUAAUA---AUAAAACUGUUUGCACGACACGCUUU .........((((((.(((((.((((((((((.((((((((((......))))((.((((...)))).)).)))))))))))---..)))))...))))).).)))))... ( -30.00, z-score = -2.08, R) >droYak2.chr3L 1307229 108 - 24197627 UUAUGCUCUGCGUGCAUGCAACGUUUUUGUUACUGUUUCUUGAUUUUCCUCGAGCGUUGCAAGGUAAAGCAGAAACAUAAUA---AUAAAACCGAUUGCACGACACGCUUU .........((((((.(((((((.((((((((.((((((((((......))))((.((((...)))).)).))))))...))---)))))).)).))))).).)))))... ( -29.80, z-score = -2.12, R) >consensus UUGUGCUCUGCGUGCAUGCAAUGUUUUUGUUACUGUUUCUCGAUUUCCUUCGAGCGUUGCAAGGUAAAGCAGAAACAUAAUAACCAUAAAACGGUUUGCACGACACGCUUU .........((((((.(((((.((((((((((.((((((((((......))))((.((((...)))).)).))))))))))).....)))))...))))).).)))))... (-28.32 = -28.16 + -0.16)

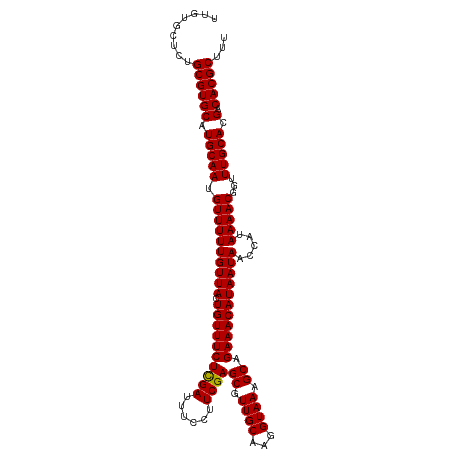
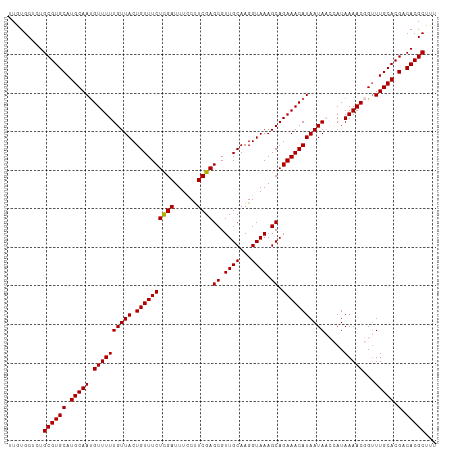
| Location | 1,348,214 – 1,348,325 |
|---|---|
| Length | 111 |
| Sequences | 5 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 94.31 |
| Shannon entropy | 0.09534 |
| G+C content | 0.39525 |
| Mean single sequence MFE | -24.92 |
| Consensus MFE | -23.64 |
| Energy contribution | -23.08 |
| Covariance contribution | -0.56 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.23 |
| Structure conservation index | 0.95 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.44 |
| SVM RNA-class probability | 0.696332 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 1348214 111 - 24543557 CUUUAUCCAUCGUUGCAAUUGUGCUCUGCGUGCAUGCAAUGUUUUUGUUACUGUUUCUCGAUUUCCUUCGAGCGUUGCAAGGUAAAGCAGAAACAUAAUAACCAUAAAACG ..........(((((((..((..(.....)..)))))))))...((((((.((((((((((......))))((.((((...)))).)).)))))))))))).......... ( -25.50, z-score = -1.30, R) >droSim1.chr3L 947199 111 - 22553184 CUUUAUCCAUCGUUGCAAUUGUGCUCUGCGUGCAUGCAAUGUUUUUGUUACUGUUUCUCGAUUUCCUUCGAGCGUUGCAAGGUAAAGCAGAAACAUAAUAACCAUAAAACG ..........(((((((..((..(.....)..)))))))))...((((((.((((((((((......))))((.((((...)))).)).)))))))))))).......... ( -25.50, z-score = -1.30, R) >droSec1.super_2 1390753 111 - 7591821 CUUUAUCCAUCGUUGCAAUUGUGCUCUGCGUGCAUGCAAUGUUUUUGUUACUGUUUCUCGAUUUCCUUCGAGCGUUGCAAGGUAAAGCAGAAACAUAAUAACCAUAAAACG ..........(((((((..((..(.....)..)))))))))...((((((.((((((((((......))))((.((((...)))).)).)))))))))))).......... ( -25.50, z-score = -1.30, R) >droEre2.scaffold_4784 1330994 108 - 25762168 CUUCAUCCAUCGCUGCAAUUAUGCUCUGCGUGCAUGCAAUGUUUUUGUUACUGUUUCUCGAUUUUCCUCGAGCGUUGCAAGGUAAAGCAGAAACAUAAUA---AUAAAACU ...(((.((.(((.(((....)))...))))).)))..((((((((((((((((..(((((......)))))....))..))))..))))))))))....---........ ( -24.20, z-score = -1.17, R) >droYak2.chr3L 1307247 108 - 24197627 CUUCAUCCAUCGCUGCAAUUAUGCUCUGCGUGCAUGCAACGUUUUUGUUACUGUUUCUUGAUUUUCCUCGAGCGUUGCAAGGUAAAGCAGAAACAUAAUA---AUAAAACC .................((((((.(((((.(((.((((((((((..(..(..(((....))).)..)..))))))))))..)))..)))))..)))))).---........ ( -23.90, z-score = -1.08, R) >consensus CUUUAUCCAUCGUUGCAAUUGUGCUCUGCGUGCAUGCAAUGUUUUUGUUACUGUUUCUCGAUUUCCUUCGAGCGUUGCAAGGUAAAGCAGAAACAUAAUAACCAUAAAACG .................((((((.(((((.(((.(((((((...............(((((......))))))))))))..)))..)))))..))))))............ (-23.64 = -23.08 + -0.56)

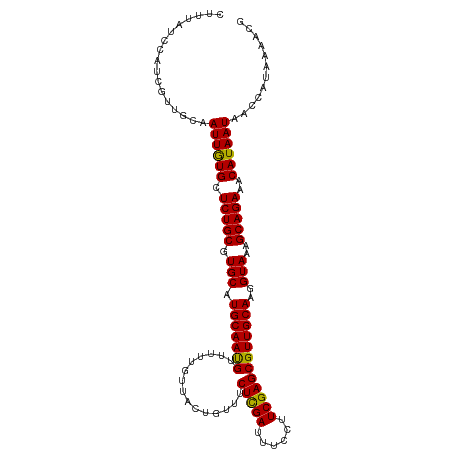
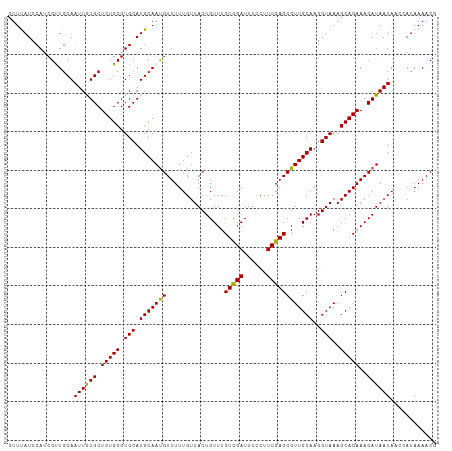
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:55:18 2011