
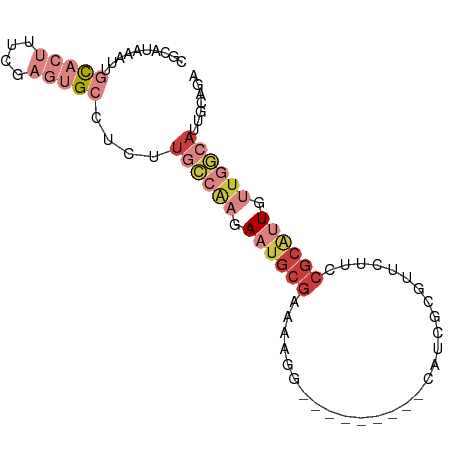
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 1,285,211 – 1,285,291 |
| Length | 80 |
| Max. P | 0.932512 |

| Location | 1,285,211 – 1,285,291 |
|---|---|
| Length | 80 |
| Sequences | 7 |
| Columns | 89 |
| Reading direction | forward |
| Mean pairwise identity | 68.82 |
| Shannon entropy | 0.59072 |
| G+C content | 0.49176 |
| Mean single sequence MFE | -25.54 |
| Consensus MFE | -9.31 |
| Energy contribution | -11.99 |
| Covariance contribution | 2.68 |
| Combinations/Pair | 1.24 |
| Mean z-score | -1.99 |
| Structure conservation index | 0.36 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.93 |
| SVM RNA-class probability | 0.854102 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
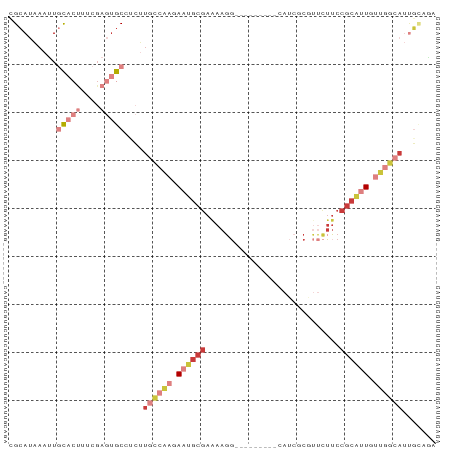
>dm3.chr3L 1285211 80 + 24543557 CGCAUAAAUUGCACUUUCGAGUGCCUCUUGCCAAGAAUGCGAAAAGG---------CAUCGCGUUCUUCCGCAAUGUUGGCAUUGCUAA .(((......(((((....)))))....))).((((((((((.....---------..))))))))))..((((((....))))))... ( -27.10, z-score = -2.45, R) >droSim1.chr3L 883781 80 + 22553184 CGCAUAAAUUGCACUUUCGAGUGCAUCUUGCCAAGAAUGCGAAAAGG---------AAUCGCCUUCUUCCGCAUUGUUGGCAUUGAUAA ..........(((((....)))))(((.((((((.((((((((((((---------.....)))).)).)))))).))))))..))).. ( -29.10, z-score = -3.89, R) >droSec1.super_2 1327779 71 + 7591821 CGCAUAAAUUGCACUUUCGAGUGCAUCUUGCCAAGAAUGCGAAAAGG---------AAUCGCGUUCUUCCGCAUUGCUAA--------- .(((.....((((((....))))))...(((.((((((((((.....---------..))))))))))..))).)))...--------- ( -24.00, z-score = -3.02, R) >droYak2.chr3L 1243657 80 + 24197627 CGCAUAAAUUGCACUUUCGAGUGCCUCUUGCCAAGAAUGCGAGAAGG---------CAUCGCGUUCUUCCGCGUUGUUGGCAUUGCAGA .(((......(((((....)))))....((((((.((((((.(((((---------........))))))))))).)))))).)))... ( -31.70, z-score = -2.87, R) >droEre2.scaffold_4784 1268156 80 + 25762168 CGCAUACAUUGCACUUUCGAGUGCCUCUUGCCAAGAAUGCGAAAAGG---------CAUCGCGUUCUUCCGCAUUGUGGGCAUUGCAGA .(((.....))).......(((((((..(((.((((((((((.....---------..))))))))))..)))....)))))))..... ( -26.70, z-score = -1.33, R) >droAna3.scaffold_13337 16659802 72 - 23293914 CGAAUAAAUU-UUCGCCCG---GCCGAGUGCCAAGAAUGCGAAAAUG---------CAACAC----UGCCGCAUUGCUGGCAUCUCAGA ((((......-))))....---...((((((((..((((((.....(---------((....----)))))))))..))))).)))... ( -21.90, z-score = -1.61, R) >droMoj3.scaffold_6680 6775021 87 + 24764193 UGCACCAACUUUUCGCCCC--UGCCGCGUGUUGACAGGCCAAAAAGUUUCUAUGGCCACGGCGCCUGUCCCAAGUGUUCAAACUGUUGA .....((((.....((...--.))(((.((..(((((((((...........))((....))))))))).)).)))........)))). ( -18.30, z-score = 1.23, R) >consensus CGCAUAAAUUGCACUUUCGAGUGCCUCUUGCCAAGAAUGCGAAAAGG_________CAUCGCGUUCUUCCGCAUUGUUGGCAUUGCAGA ..........(((((....)))))....((((((.((((((............................)))))).))))))....... ( -9.31 = -11.99 + 2.68)

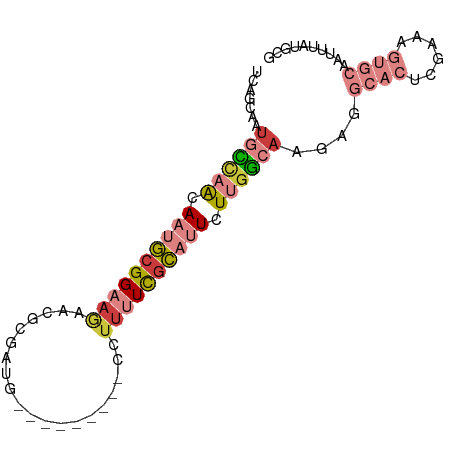
| Location | 1,285,211 – 1,285,291 |
|---|---|
| Length | 80 |
| Sequences | 7 |
| Columns | 89 |
| Reading direction | reverse |
| Mean pairwise identity | 68.82 |
| Shannon entropy | 0.59072 |
| G+C content | 0.49176 |
| Mean single sequence MFE | -25.31 |
| Consensus MFE | -11.35 |
| Energy contribution | -13.54 |
| Covariance contribution | 2.19 |
| Combinations/Pair | 1.35 |
| Mean z-score | -1.73 |
| Structure conservation index | 0.45 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.37 |
| SVM RNA-class probability | 0.932512 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
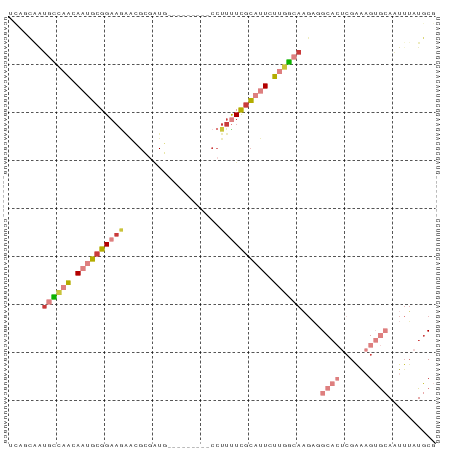
>dm3.chr3L 1285211 80 - 24543557 UUAGCAAUGCCAACAUUGCGGAAGAACGCGAUG---------CCUUUUCGCAUUCUUGGCAAGAGGCACUCGAAAGUGCAAUUUAUGCG ...(((.((((((...((((((((...((...)---------).))))))))...))))))....(((((....)))))......))). ( -25.70, z-score = -1.96, R) >droSim1.chr3L 883781 80 - 22553184 UUAUCAAUGCCAACAAUGCGGAAGAAGGCGAUU---------CCUUUUCGCAUUCUUGGCAAGAUGCACUCGAAAGUGCAAUUUAUGCG .......((((((.(((((((((..(((.....---------)))))))))))).))))))...((((((....))))))......... ( -27.90, z-score = -3.55, R) >droSec1.super_2 1327779 71 - 7591821 ---------UUAGCAAUGCGGAAGAACGCGAUU---------CCUUUUCGCAUUCUUGGCAAGAUGCACUCGAAAGUGCAAUUUAUGCG ---------...(((.(((.((.(((.((((..---------.....)))).))))).)))...((((((....)))))).....))). ( -22.80, z-score = -2.33, R) >droYak2.chr3L 1243657 80 - 24197627 UCUGCAAUGCCAACAACGCGGAAGAACGCGAUG---------CCUUCUCGCAUUCUUGGCAAGAGGCACUCGAAAGUGCAAUUUAUGCG ...(((.((((((...((((......))))(((---------(......))))..))))))....(((((....)))))......))). ( -26.10, z-score = -1.90, R) >droEre2.scaffold_4784 1268156 80 - 25762168 UCUGCAAUGCCCACAAUGCGGAAGAACGCGAUG---------CCUUUUCGCAUUCUUGGCAAGAGGCACUCGAAAGUGCAAUGUAUGCG ((((((.((....)).))))))...((.(((((---------((((((.((.......)))))))))).)))...))(((.....))). ( -24.70, z-score = -1.02, R) >droAna3.scaffold_13337 16659802 72 + 23293914 UCUGAGAUGCCAGCAAUGCGGCA----GUGUUG---------CAUUUUCGCAUUCUUGGCACUCGGC---CGGGCGAA-AAUUUAUUCG .(((((.((((((.(((((((.(----(((...---------)))).))))))).))))))))))).---....((((-......)))) ( -25.60, z-score = -1.96, R) >droMoj3.scaffold_6680 6775021 87 - 24764193 UCAACAGUUUGAACACUUGGGACAGGCGCCGUGGCCAUAGAAACUUUUUGGCCUGUCAACACGCGGCA--GGGGCGAAAAGUUGGUGCA (((((..((((..(.((((...)))).((((((((((...........)))))((....)).))))).--.)..))))..))))).... ( -24.40, z-score = 0.63, R) >consensus UCAGCAAUGCCAACAAUGCGGAAGAACGCGAUG_________CCUUUUCGCAUUCUUGGCAAGAGGCACUCGAAAGUGCAAUUUAUGCG .......((((((.((((((((((....................)))))))))).))))))....((((......)))).......... (-11.35 = -13.54 + 2.19)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:55:13 2011