
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 1,255,214 – 1,255,318 |
| Length | 104 |
| Max. P | 0.761562 |

| Location | 1,255,214 – 1,255,318 |
|---|---|
| Length | 104 |
| Sequences | 7 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 72.64 |
| Shannon entropy | 0.50730 |
| G+C content | 0.53119 |
| Mean single sequence MFE | -36.46 |
| Consensus MFE | -17.54 |
| Energy contribution | -19.09 |
| Covariance contribution | 1.54 |
| Combinations/Pair | 1.35 |
| Mean z-score | -1.61 |
| Structure conservation index | 0.48 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.61 |
| SVM RNA-class probability | 0.761562 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
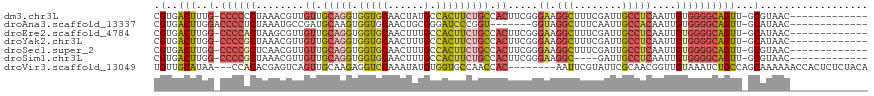
>dm3.chr3L 1255214 104 + 24543557 CGUGACUUUG-CCCCCCUAAACGUUGUUGCAGGUGGUGCAACUAUGCCACUUCUGCCACUUCGGGAAGGCUUUCGAUUGCCUCAAUUGUGGGGCAGUU-GCGUAAC------------- ((..(((..(-((((......((..((.(((((((((........)))))..)))).))..)).((.(((........)))))......)))))))).-.))....------------- ( -37.60, z-score = -1.83, R) >droAna3.scaffold_13337 16628919 98 - 23293914 CGUGACUUGGACCCCUCUAAAUGCCGAUGCAAGUGGUGCAACUGCGGGAUCCCGGU-------GGUAGGCUUUCAAUUGCCACAAUUGUGGGGCAGUU-GCAUAAC------------- ......(((((....))))).(((....))).((.((((((((((((....)).((-------(((((........))))))).........))))))-)))).))------------- ( -31.50, z-score = -0.11, R) >droEre2.scaffold_4784 1238539 104 + 25762168 CGUGACUUGG-CCCCACUAAGCGUUGUUGCAGGUGGUGCAACUUUGCCACUUCUGCCACUUCGGGAAGGCUUUCGAUUGCCUCAAUUGUGGGGCAGUU-GCAUAAC------------- .(..(((..(-((((((....((..((.(((((((((........)))))..)))).))..)).((.(((........)))))....)))))))))).-.).....------------- ( -41.00, z-score = -2.36, R) >droYak2.chr3L 1213200 104 + 24197627 CGUGACUUGG-CCCCGCUAAACGUUGUUGCAGGUGGUGCAACUUUGCCACUUCUGCCACUUCGGGAAGGCUUUCGAUUGCCUCAAUUGUGGGGCAGUU-GCAUAAC------------- .(..(((..(-((((((....((..((.(((((((((........)))))..)))).))..)).((.(((........)))))....)))))))))).-.).....------------- ( -40.90, z-score = -2.51, R) >droSec1.super_2 1297886 104 + 7591821 CGUGACUUGG-CCCCGCUCAACGUUGUUGCAGGUGGUGCAACUUUGCCACUUCUGCCACUUCGGGAAGGCUUUCGAUUGCCUCAAUUGUGGGGCAGUU-GCGUAAC------------- ((..(((..(-((((((....((..((.(((((((((........)))))..)))).))..)).((.(((........)))))....)))))))))).-.))....------------- ( -41.90, z-score = -2.42, R) >droSim1.chr3L 853059 100 + 22553184 CGUGACUUGG-CCCCGCUAAACGUUGUUGCAGGUGGUGCAACUUUGCCACUUCUGCCACUUCGGGAAGGC----GAUUGCCUCAAUUGUGGGGCAGUU-GCGUAAC------------- ((..(((..(-((((((....((..((.(((((((((........)))))..)))).))..)).((.(((----....)))))....)))))))))).-.))....------------- ( -41.00, z-score = -2.31, R) >droVir3.scaffold_13049 16631207 108 - 25233164 UGUUGCAUAA---CCACACGAGUCAGUUGCAAGAGGUCCAAAUAUGUGGUGCCAACCAC--------AAUUCGUAUUCGCAACGGUUGUAAAUCUGCCAGCAAAAAACCACUCUCUACA ..((((....---............(((((..(((...(.(((.((((((....)))))--------)))).)..))))))))(((.(.....).))).))))................ ( -21.30, z-score = 0.24, R) >consensus CGUGACUUGG_CCCCGCUAAACGUUGUUGCAGGUGGUGCAACUUUGCCACUUCUGCCACUUCGGGAAGGCUUUCGAUUGCCUCAAUUGUGGGGCAGUU_GCAUAAC_____________ .((((((....((((((....((..((.(((((.(((((......).))))))))).))..)).((.(((........)))))....)))))).)))).)).................. (-17.54 = -19.09 + 1.54)


Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:55:11 2011