
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 1,219,871 – 1,219,988 |
| Length | 117 |
| Max. P | 0.998643 |

| Location | 1,219,871 – 1,219,988 |
|---|---|
| Length | 117 |
| Sequences | 6 |
| Columns | 129 |
| Reading direction | forward |
| Mean pairwise identity | 57.09 |
| Shannon entropy | 0.76122 |
| G+C content | 0.46509 |
| Mean single sequence MFE | -33.27 |
| Consensus MFE | -19.58 |
| Energy contribution | -19.72 |
| Covariance contribution | 0.14 |
| Combinations/Pair | 1.27 |
| Mean z-score | -1.61 |
| Structure conservation index | 0.59 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.43 |
| SVM RNA-class probability | 0.998643 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
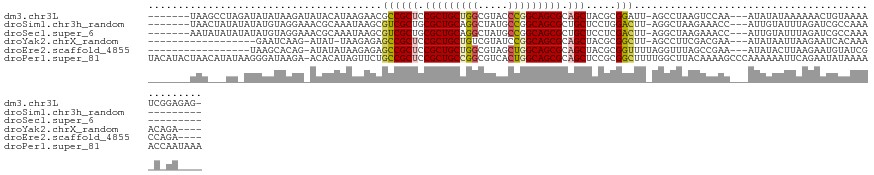
>dm3.chr3L 1219871 117 + 24543557 -------UAAGCCUAGAUAUAUAAGAUAUACAUAAGAACGCCGCUCCGCUGCUGGCGUACCCGGCAGCGCAGCUACGCGGAUU-AGCCUAAGUCCAA---AUAUAUAAAAAACUGUAAAAUCGGAGAG- -------..........((((((................((.(((.(((((((((.....))))))))).)))...))(((((-......)))))..---.)))))).....(((......)))....- ( -31.10, z-score = -1.46, R) >droSim1.chr3h_random 862046 109 + 1452968 -------UAACUAUAUAUAUGUAGGAAACGCAAAUAAGCGUCGCUGCGCUGCAGGCUAUGCCGGCAGCGCUGCUCCUGGACUU-AGGCUAAGAAACC---AUUGUAUUUAGAUCGCCAAA--------- -------...(((.((((((.(((((.((((......)))).((.(((((((.(((...))).))))))).))))))).)...-.((........))---..))))).))).........--------- ( -35.90, z-score = -1.65, R) >droSec1.super_6 688995 109 - 4358794 -------AAUAUAUAUAUAUGUAGGAAACGCAAAUAAGCGUCGCUGCGCUGCAGGCUAUGCCGGCAGCGCUGCUCCUCGACUU-AGGCUAAGAAACC---AUUGUAUUUAGAUCGCCAAA--------- -------............((.((((.((((......)))).((.(((((((.(((...))).))))))).))))))))....-.(((.........---..............)))...--------- ( -32.70, z-score = -0.95, R) >droYak2.chrX_random 1263049 101 - 1802292 ------------------GAAUCAAG-AUAU-UAAGAGAGCCGCUCCGCUGCUGUCGUAUCCGGCAGCGCAGCUACGCGGCUU-AGCCUUCGACGAA---AUAUAAUUAAGAAUCACAAAACAGA---- ------------------((.((...-....-.....(((((((...((((((((((....)))))..)))))...)))))))-....(((...)))---..........)).))..........---- ( -26.10, z-score = -0.78, R) >droEre2.scaffold_4855 234970 104 - 242605 -----------------UAAGCACAG-AUAUAUAAGAGAGCCGCUCCGCUGCUGGCGUAGCUGGCAGCGCAGCUACGCGGUUUUAGGUUUAGCCGAA---AUAUACUUAAGAAUGUAUCGCCAGA---- -----------------........(-((((((...((((((((..((((((..((...))..)))))).......)))))))).((.....))...---............)))))))......---- ( -33.00, z-score = -0.63, R) >droPer1.super_81 10280 128 + 277874 UACAUACUAACAUAUAAGGGAUAAGA-ACACAUAGUUCUGCCGCUCCGCUGCCGGCGUCACUGGCAGCGCAGCUCCGCGGCUUUUGGCUUACAAAAGCCCAAAAAAUUCAGAAUAUAAAAACCAAUAAA ..................((...(((-((.....))))).))(((.(((((((((.....))))))))).)))...(.((((((((.....)))))))))............................. ( -40.80, z-score = -4.21, R) >consensus _______UAA___UA_AUAUAUAAGA_ACACAUAAGAGCGCCGCUCCGCUGCUGGCGUAGCCGGCAGCGCAGCUACGCGGCUU_AGGCUAAGAAAAA___AUAUAAUUAAGAAUGUAAAA_C_GA____ .......................................((((((.(((((((((.....))))))))).))).....)))................................................ (-19.58 = -19.72 + 0.14)

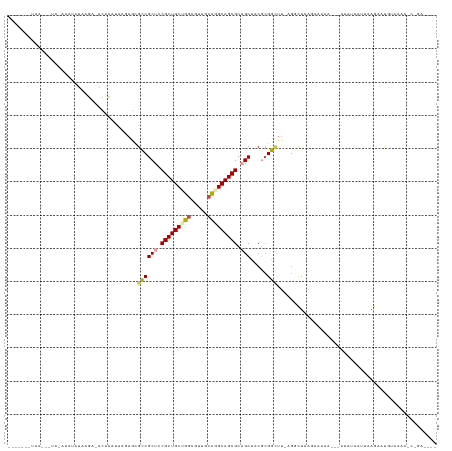
| Location | 1,219,871 – 1,219,988 |
|---|---|
| Length | 117 |
| Sequences | 6 |
| Columns | 129 |
| Reading direction | reverse |
| Mean pairwise identity | 57.09 |
| Shannon entropy | 0.76122 |
| G+C content | 0.46509 |
| Mean single sequence MFE | -33.41 |
| Consensus MFE | -17.70 |
| Energy contribution | -18.53 |
| Covariance contribution | 0.83 |
| Combinations/Pair | 1.00 |
| Mean z-score | -0.82 |
| Structure conservation index | 0.53 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.65 |
| SVM RNA-class probability | 0.958110 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
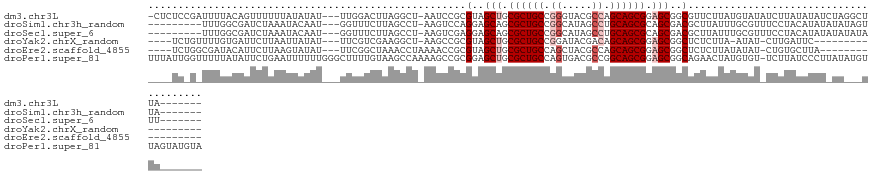
>dm3.chr3L 1219871 117 - 24543557 -CUCUCCGAUUUUACAGUUUUUUAUAUAU---UUGGACUUAGGCU-AAUCCGCGUAGCUGCGCUGCCGGGUACGCCAGCAGCGGAGCGGCGUUCUUAUGUAUAUCUUAUAUAUCUAGGCUUA------- -...(((((....................---)))))..((((((-.....((((.(((.((((((.((.....)).)))))).))).))))...((((((.....))))))....))))))------- ( -34.15, z-score = -0.79, R) >droSim1.chr3h_random 862046 109 - 1452968 ---------UUUGGCGAUCUAAAUACAAU---GGUUUCUUAGCCU-AAGUCCAGGAGCAGCGCUGCCGGCAUAGCCUGCAGCGCAGCGACGCUUAUUUGCGUUUCCUACAUAUAUAUAGUUA------- ---------...(((.....((((.....---.))))....))).-......((((((.(((((((.(((...))).))))))).))(((((......)))))))))...............------- ( -35.40, z-score = -1.63, R) >droSec1.super_6 688995 109 + 4358794 ---------UUUGGCGAUCUAAAUACAAU---GGUUUCUUAGCCU-AAGUCGAGGAGCAGCGCUGCCGGCAUAGCCUGCAGCGCAGCGACGCUUAUUUGCGUUUCCUACAUAUAUAUAUAUU------- ---------....((..............---.((((((..((..-..))..)))))).(((((((.(((...))).))))))).))(((((......)))))...................------- ( -35.60, z-score = -1.80, R) >droYak2.chrX_random 1263049 101 + 1802292 ----UCUGUUUUGUGAUUCUUAAUUAUAU---UUCGUCGAAGGCU-AAGCCGCGUAGCUGCGCUGCCGGAUACGACAGCAGCGGAGCGGCUCUCUUA-AUAU-CUUGAUUC------------------ ----...(...((((((.....)))))).---..)....((((..-.((((((....((((((((.((....)).)))).)))).)))))).)))).-....-........------------------ ( -27.30, z-score = 0.13, R) >droEre2.scaffold_4855 234970 104 + 242605 ----UCUGGCGAUACAUUCUUAAGUAUAU---UUCGGCUAAACCUAAAACCGCGUAGCUGCGCUGCCAGCUACGCCAGCAGCGGAGCGGCUCUCUUAUAUAU-CUGUGCUUA----------------- ----................((((((((.---...((.....)).......((...(((.((((((..((...))..)))))).))).))............-.))))))))----------------- ( -25.60, z-score = 1.17, R) >droPer1.super_81 10280 128 - 277874 UUUAUUGGUUUUUAUAUUCUGAAUUUUUUGGGCUUUUGUAAGCCAAAAGCCGCGGAGCUGCGCUGCCAGUGACGCCGGCAGCGGAGCGGCAGAACUAUGUGU-UCUUAUCCCUUAUAUGUUAGUAUGUA ............(((((.((((........((((((((.....))))))))..((((((.(((((((.(.....).))))))).)))...(((((.....))-)))..)))........)))).))))) ( -42.40, z-score = -2.01, R) >consensus ____UC_G_UUUGACAUUCUUAAUUAUAU___UUUGUCUAAGCCU_AAGCCGCGGAGCUGCGCUGCCGGCUACGCCAGCAGCGGAGCGGCGCUCUUAUGUAU_UCUUACAUAU_UA___UUA_______ .....................................................(..(((.((((((.((.....)).)))))).)))..)....................................... (-17.70 = -18.53 + 0.83)


Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:55:10 2011