| Sequence ID | dm3.chr2L |
|---|---|
| Location | 4,094,228 – 4,094,318 |
| Length | 90 |
| Max. P | 0.878779 |

| Location | 4,094,228 – 4,094,318 |
|---|---|
| Length | 90 |
| Sequences | 10 |
| Columns | 94 |
| Reading direction | forward |
| Mean pairwise identity | 73.10 |
| Shannon entropy | 0.55625 |
| G+C content | 0.36773 |
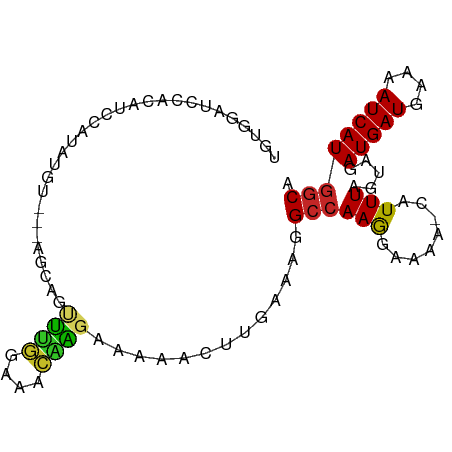
| Mean single sequence MFE | -16.77 |
| Consensus MFE | -8.44 |
| Energy contribution | -7.96 |
| Covariance contribution | -0.48 |
| Combinations/Pair | 1.50 |
| Mean z-score | -1.49 |
| Structure conservation index | 0.50 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.04 |
| SVM RNA-class probability | 0.878779 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
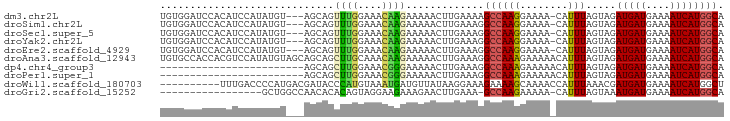
>dm3.chr2L 4094228 90 + 23011544 UGUGGAUCCACAUCCAUAUGU---AGCAGUUUGGAAACAAGAAAAACUUGAAAAGCCAAGGAAAA-CAUUUAGUAGAUGAUGAAAAUCAUGGCA (((((((....)))))))(((---.....((((....)))).....((((......))))....)-))....((..(((((....))))).)). ( -19.50, z-score = -1.85, R) >droSim1.chr2L 4049276 90 + 22036055 UGUGGAUCCACAUCCAUAUGU---AGCAGUUUGGAAACAAGAAAAACUUGAAAGGCCAAGGAAAA-CAUUUAGUAGAUGAUGAAAAUCAUGGCA (((((((....)))))))(((---.....((((....)))).....((((......))))....)-))....((..(((((....))))).)). ( -19.50, z-score = -1.49, R) >droSec1.super_5 2193262 90 + 5866729 UGUGGAUCCACAUCCAUAUGU---AGCAGUUUGGAAACAAGAAAAACUUGAAAGGCCAAGGAAAA-CAUUUAGUAGAUGAUGAAAAUCAUGGCA (((((((....)))))))(((---.....((((....)))).....((((......))))....)-))....((..(((((....))))).)). ( -19.50, z-score = -1.49, R) >droYak2.chr2L 4111410 90 + 22324452 UGUGGAUCCACAUCCAUAUGU---AGCAGUUUGGAAACAAGAAAAACUUGAAAGGCCAAGGAAAA-CAUUUAGUAGAUGAUGAAAAUCAUGGCA (((((((....)))))))(((---.....((((....)))).....((((......))))....)-))....((..(((((....))))).)). ( -19.50, z-score = -1.49, R) >droEre2.scaffold_4929 4152236 90 + 26641161 UGUGGAUCCACAUCCAUAUGU---AGCAGUUUGGAAACAAGAAAAACUUGAAAGGCCAAGGAAAA-CAUUUAGUAGAUGAUGAAAAUCAUGGCA (((((((....)))))))(((---.....((((....)))).....((((......))))....)-))....((..(((((....))))).)). ( -19.50, z-score = -1.49, R) >droAna3.scaffold_12943 660068 94 - 5039921 UGUGCCACCACGUCCAUAUGUAGCAGCAGCUUGCAAACAAGAAAAACUUGAAAGGCCAAAGAAAAACAUUUAGUAGAUGAUGAAAAUCAUGGCA ..((((.....(.((...((((((....)).))))..((((.....))))...)).)...................(((((....))))))))) ( -18.40, z-score = -0.65, R) >dp4.chr4_group3 9846845 70 - 11692001 ------------------------AGCAGCUUGGAAACGGGAAAAACUUGAAAGGCCAAAGAAAAACAUUUAGUAGAUGAUGAAAAUCAUGGCA ------------------------.....((((....)))).............(((........((.....))..(((((....)))))))). ( -13.80, z-score = -2.01, R) >droPer1.super_1 6966669 70 - 10282868 ------------------------AGCAGCUUGGAAACGGGAAAAACUUGAAAGGCCAAAGAAAAACAUUUAGUAGAUGAUGAAAAUCAUGGCA ------------------------.....((((....)))).............(((........((.....))..(((((....)))))))). ( -13.80, z-score = -2.01, R) >droWil1.scaffold_180703 2090543 84 + 3946847 ----------UUUGACCCCAUGACGAUACCCAUGUAAAUGAUGUUAUAAGGAAAGAAAAGCAAAACCAUUUAAACGAUGAUGAAAAUCAUGGCU ----------.......((((((......((.(((((......))))).))...............((((........))))....)))))).. ( -11.60, z-score = -1.21, R) >droGri2.scaffold_15252 12677944 75 + 17193109 -----------------GCUGGCCAACACACAGUAGGAAGAAAGAACUUGAAA-GCCAAGAAAAA-CAUUUAGUAAAUGAUGAAAAUCAUGGCA -----------------....(((......................((((...-..)))).....-..........(((((....)))))))). ( -12.60, z-score = -1.22, R) >consensus UGUGGAUCCACAUCCAUAUGU___AGCAGUUUGGAAACAAGAAAAACUUGAAAGGCCAAGGAAAA_CAUUUAGUAGAUGAUGAAAAUCAUGGCA .............................((((....)))).............(((.........((((.....))))(((.....)))))). ( -8.44 = -7.96 + -0.48)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:15:22 2011