| Sequence ID | dm3.chr3L |
|---|---|
| Location | 1,212,786 – 1,212,886 |
| Length | 100 |
| Max. P | 0.957261 |

| Location | 1,212,786 – 1,212,886 |
|---|---|
| Length | 100 |
| Sequences | 8 |
| Columns | 103 |
| Reading direction | reverse |
| Mean pairwise identity | 59.36 |
| Shannon entropy | 0.78395 |
| G+C content | 0.50372 |
| Mean single sequence MFE | -26.75 |
| Consensus MFE | -13.05 |
| Energy contribution | -12.23 |
| Covariance contribution | -0.81 |
| Combinations/Pair | 1.73 |
| Mean z-score | -0.92 |
| Structure conservation index | 0.49 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.64 |
| SVM RNA-class probability | 0.957261 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
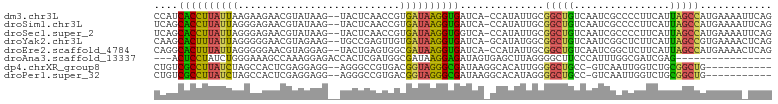
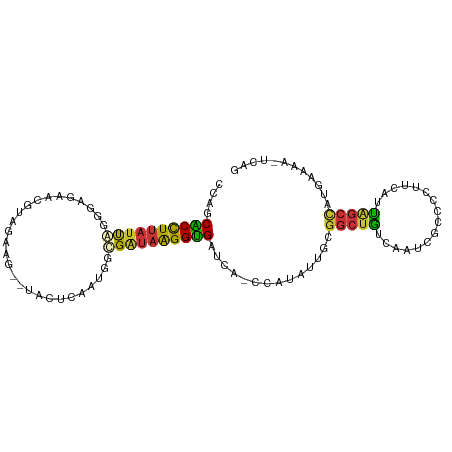
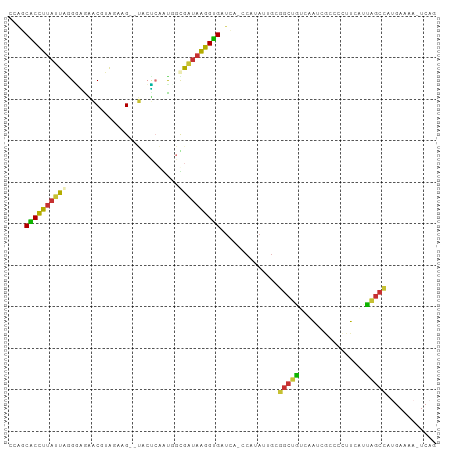
>dm3.chr3L 1212786 100 - 24543557 CCAUCACCUUAUUAAGAAGAACGUAUAAG--UACUCAACCGUGAUAAGGUGAUCA-CCAUAUUGCGGCUGUCAAUCGCCCCUUCAUUAGCCAUGAAAAUUCAG ..((((((((((((...((.((......)--).))......))))))))))))..-.........(((((................)))))............ ( -20.39, z-score = -1.99, R) >droSim1.chr3L 829132 100 - 22553184 UCAGCACCUUAUUAGGGAGAACGUAUAAG--UACUCAACCGUGAUAAGGUGAUCA-CCAUAUUGCGGCUGUCAAUCGCCCCUUCAUUAGCCAUGAAAAUUCAG (((.(((((((((((((((.((......)--).)))..)).))))))))))....-.........(((((................))))).)))........ ( -24.49, z-score = -1.85, R) >droSec1.super_2 1206613 100 - 7591821 UCAGCACCUUAUUAGGGAGAACGUAUAAG--UACUCAACCGUGAUAAGGUGGUCA-CCAUAUUGCGGCUGUCAAUCGCCCCUUCAUUAGCCAUGAAAAUUCAG (((.(((((((((((((((.((......)--).)))..)).))))))))))....-.........(((((................))))).)))........ ( -24.69, z-score = -1.44, R) >droYak2.chr3L 1178858 100 - 24197627 CAAGCACUUUAUUAGGGGGAACGUAGAAG--UGCCGAGUUGUGAUAAGGUGAUCA-GCAUAUGGCGGCUGUCAAUCGGCUCUUCAUUAGCCGUGAAAACUCAG ...(((((((..((.(.....).))))))--))).(((((.........(((.((-((........)))))))..(((((.......)))))....))))).. ( -26.60, z-score = -0.46, R) >droEre2.scaffold_4784 1202802 100 - 25762168 CAGGCACUUUAUUAGGGGGAACGUAGGAG--UACUGAGUGGCGAUAAGGUGAUCA-CCAUAUUGCGGCUGUCAAUCGGCUCUUCAUUAGCCAUGAAAACUCAG ..(((...........(....)(.(((((--(..(((((.((((((.(((....)-)).)))))).))..)))....)))))))....)))............ ( -26.10, z-score = -0.57, R) >droAna3.scaffold_13337 6701208 84 - 23293914 ---ACUCCUAUCUGGGAAAGCCAAAGGAGACCACUCGAUGGCGAUAAGGAGAUAGUGAGCUUAGGGGCUUCCCAUUUGGCGAUCGAG---------------- ---.(((((...(((.....))).)))))....((((((.((.(...((.((.....((((....))))))))...).)).))))))---------------- ( -25.70, z-score = -0.57, R) >dp4.chrXR_group8 8303665 89 + 9212921 CUGUCGCCUUAUCUAGCCACUCGAGGAGG--AGGGCCGUGACGGUAGGGCGAUAAGGCACAUUGGGGCUGCC-GUCAAUUGGUCUGCGGCUG----------- .(((.((((((((..(((.(((......)--)).((((...))))..))))))))))))))....((((((.-(.(.....).).)))))).----------- ( -33.00, z-score = -0.22, R) >droPer1.super_32 572106 89 - 992631 CUGUCGCCUUAUCUAGCCACUCGAGGAGG--AGGGCCGUGACGGUAGGGCGAUAAGGCACAUAGGGGCUGCC-GUCAAUUGGUCUGCGGCUG----------- .(((.((((((((..(((.(((......)--)).((((...))))..))))))))))))))....((((((.-(.(.....).).)))))).----------- ( -33.00, z-score = -0.25, R) >consensus CCAGCACCUUAUUAGGGAGAACGUAGAAG__UACUCAAUGGCGAUAAGGUGAUCA_CCAUAUUGCGGCUGUCAAUCGCCCCUUCAUUAGCCAUGAAAA_UCAG ....((((((((((...........................))))))))))..............(((((................)))))............ (-13.05 = -12.23 + -0.81)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:55:09 2011