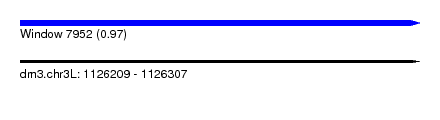
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 1,126,209 – 1,126,307 |
| Length | 98 |
| Max. P | 0.973439 |

| Location | 1,126,209 – 1,126,307 |
|---|---|
| Length | 98 |
| Sequences | 10 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 84.77 |
| Shannon entropy | 0.29865 |
| G+C content | 0.32003 |
| Mean single sequence MFE | -25.38 |
| Consensus MFE | -8.20 |
| Energy contribution | -8.51 |
| Covariance contribution | 0.31 |
| Combinations/Pair | 1.19 |
| Mean z-score | -4.11 |
| Structure conservation index | 0.32 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.89 |
| SVM RNA-class probability | 0.973439 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
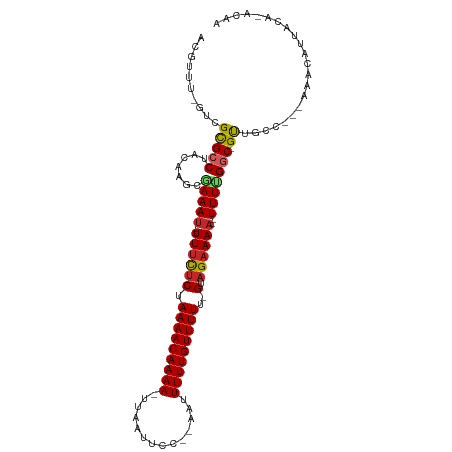
>dm3.chr3L 1126209 98 + 24543557 ACGUUU-GUCGCGCCUACAAGCGAAAU--UUUCUCUAAAACAAAA-UUAAUUCC--AAUUUUUGUUUUU-GUAGAAAA--UUUUGGC-GUUGGC---AAACAUUACA-ACAA ..((((-((((((((.......(((((--((((((.(((((((((-........--...))))))))).-).))))))--)))))))-).))))---))))......-.... ( -29.31, z-score = -4.92, R) >droSim1.chr3L 754192 98 + 22553184 ACGUUU-GUCGCGCCUACGAGCGAAAU--UUUCUCUAAAACAAAA-UUAAUUCC--AAUUUUUGUUUUU-GUAGAAAA--UUUUGGC-GUUGGC---AAACAUUACA-ACAA ..((((-((((((((.......(((((--((((((.(((((((((-........--...))))))))).-).))))))--)))))))-).))))---))))......-.... ( -29.31, z-score = -4.54, R) >droSec1.super_2 1128534 92 + 7591821 -------AUCGCGCCUACGAGCGAAAU--UUUCUCUAAAACAAAA-UUAAUUCC--AAUUUUUGUUUUU-GUAGAAAA--UUUUGGC-GUUGGC---AAACAUUACA-ACAA -------.....(((.(((..((((((--((((((.(((((((((-........--...))))))))).-).))))))--))))).)-)).)))---..........-.... ( -25.90, z-score = -4.52, R) >droYak2.chr3L 1097335 98 + 24197627 ACGUUU-GUCGCGCCUACAAGCGAAAU--UUUCUCUAAAACAAAA-UUAAUUCC--AAUUUUUGUUUUU-GUAGAAAA--UUUCGGC-GCUGGC---AAACAUUACA-ACAA ..((((-((((((((.......(((((--((((((.(((((((((-........--...))))))))).-).))))))--)))))))-)).)))---))))......-.... ( -32.81, z-score = -6.37, R) >droEre2.scaffold_4784 1126867 98 + 25762168 ACGUUU-GUCGCGCCUACAAGCGAAAU--UUUCUCUAAAACAAAA-UUAAUUCC--AAUUUUUGUUUUU-GUAGAAAA--UUUUGGC-GUUGCC---AAACAUUACA-ACAA ..((((-(.((((((.......(((((--((((((.(((((((((-........--...))))))))).-).))))))--)))))))-).)).)---))))......-.... ( -24.91, z-score = -3.85, R) >droPer1.super_45 6365 111 - 618639 ACGUUUUGUCGCGCCUACAAGCGAAAUAUUUCUCCUAAAACAAAAAUUAAUUUCCAAAUUUUUGUUUUUUGUAGAAAAAUUUUUGGCCGUUGCCCCAAAACAUUUCA-ACAA ..((((((((((........))))....((((((..((((((((((((........))))))))))))..).))))).......(((....))).))))))......-.... ( -24.70, z-score = -3.80, R) >dp4.chrXR_group8 5869259 101 + 9212921 ACGUUU-GUCGCGCCUACAAGCGAAAU--UUUCUCUAAAACAAAA-UUAAUUCC--AAUUUUUGUUUUU-GUAGAAAA--UUUUGGC-GUUGCCCAAAAACAUUACA-ACAA ..((((-((.(((((.......(((((--((((((.(((((((((-........--...))))))))).-).))))))--)))))))-)).))....))))......-.... ( -22.31, z-score = -2.90, R) >droWil1.scaffold_180698 4776960 98 + 11422946 GCGUUU-GUCGCGCCUACAACCAAAAA-UUUUCUCUAAAACAAAA-UUAAUUCC--AAUUUUUGUUUUU-GUAGAAAA--UUUUGGC-GUUGCC----AACAUUACACACA- ..(((.-((.(((((........((((-(((((((.(((((((((-........--...))))))))).-).))))))--)))))))-)).)).----)))..........- ( -23.90, z-score = -4.14, R) >droMoj3.scaffold_6680 23996673 91 - 24764193 ------ACAUGCGACUGCAC---AAAU--UUUCUCUAAAACAAAA-UUAAUUCC--GAUUUUUGUUUUU-GUAGAAAA-UUUUCGGC-GCUGGCU---AACAUUACA-ACAA ------.((.(((.(((...---((((--((((((.(((((((((-........--...))))))))).-).))))))-))).))))-))))...---.........-.... ( -19.50, z-score = -3.09, R) >droVir3.scaffold_13049 421501 93 + 25233164 ------ACAUGUGACUGCACUGCAAAU--UUUCUCUAAAACAAAA-UUAAUUCC--GUAUUUUGUUUUU-GUAGAAAA-UUUU-GGC-GUUGCCC---AACAUUACA-ACAA ------..((((....(((.(((.(((--((((((.(((((((((-(.......--..)))))))))).-).))))))-))..-.))-).)))..---.))))....-.... ( -21.20, z-score = -3.01, R) >consensus ACGUUU_GUCGCGCCUACAAGCGAAAU__UUUCUCUAAAACAAAA_UUAAUUCC__AAUUUUUGUUUUU_GUAGAAAA__UUUUGGC_GUUGCC___AAACAUUACA_ACAA .....................(((((...(((((..(((((((((.((........)).)))))))))....)))))...)))))........................... ( -8.20 = -8.51 + 0.31)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:55:04 2011