| Sequence ID | dm3.chr3L |
|---|---|
| Location | 1,071,921 – 1,072,022 |
| Length | 101 |
| Max. P | 0.996439 |

| Location | 1,071,921 – 1,072,022 |
|---|---|
| Length | 101 |
| Sequences | 10 |
| Columns | 102 |
| Reading direction | forward |
| Mean pairwise identity | 72.87 |
| Shannon entropy | 0.57616 |
| G+C content | 0.53168 |
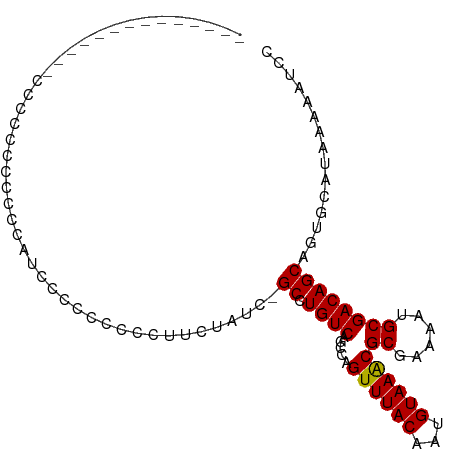
| Mean single sequence MFE | -16.90 |
| Consensus MFE | -12.63 |
| Energy contribution | -12.38 |
| Covariance contribution | -0.25 |
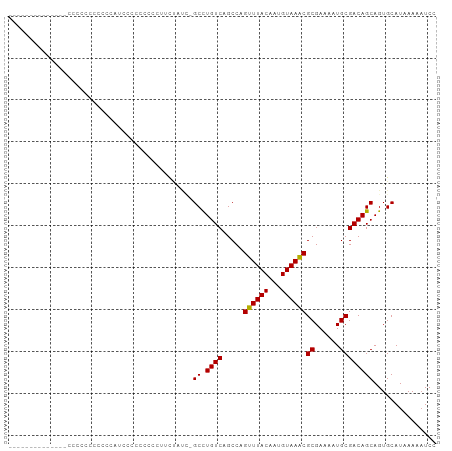
| Combinations/Pair | 1.07 |
| Mean z-score | -1.81 |
| Structure conservation index | 0.75 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.93 |
| SVM RNA-class probability | 0.996439 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
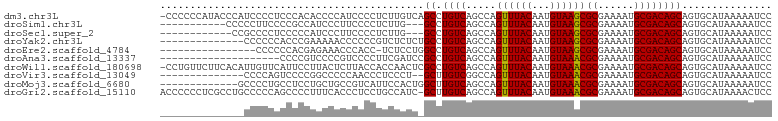
>dm3.chr3L 1071921 101 + 24543557 -CCCCCCAUACCCAUCCCCUCCCACACCCCAUCCCCUCUUGUCAGCCUGUCAGCCAGUUUACAAUGUAAGCGCGAAAAUGCGACAGCAGUGCAUAAAAAUCC -.....................(((....((........))...((.((((.....((((((...))))))((......)))))))).)))........... ( -13.30, z-score = -1.78, R) >droSim1.chr3L 698519 88 + 22553184 -----------CCCCCUUCCCCGCCAUCCCUUCCCCUCUUG---GCCUGUCAGCCAGUUUACAAUGUAAGCGCGAAAAUGCGACAGCAGUGCAUAAAAAUCC -----------...........((((.............))---))(((((.....((((((...))))))((......)))))))................ ( -15.92, z-score = -1.52, R) >droSec1.super_2 1075336 87 + 7591821 ------------CCGCCCCUCCCCCAUCCCUUCCCCUCUUG---GCCUGUCAGCCAGUUUACAAUGUAAGCGCGAAAAUGCGACAGCAGUGCAUAAAAAUCC ------------.(((.......................((---((......))))((((((...)))))))))...((((.((....))))))........ ( -15.80, z-score = -1.46, R) >droYak2.chr3L 1041923 88 + 24197627 --------------CCCCCCACCCGAAAAACCCCCCGUCUCUCUGCCUGUCAGCCAGUUUACAAUGUAAGCGCGAAAAUGCGACAGCAGUGCAUAAAAAUCC --------------......................((....((((.((((.....((((((...))))))((......)))))))))).)).......... ( -16.00, z-score = -2.66, R) >droEre2.scaffold_4784 1069315 85 + 25762168 ----------------CCCCCCACGAGAAACCCACC-UCUCCUGGCCUGUCAGCCAGUUUACAAUGUAAGCGCGAAAAUGCGACAGCAGUGCAUAAAAAUCC ----------------....(((.((((........-)))).))).(((((.....((((((...))))))((......)))))))................ ( -18.50, z-score = -2.19, R) >droAna3.scaffold_13337 16448312 82 - 23293914 --------------------CCCCGUCCCCGUCCCCUUCGAUCCGCCUGUCAGCCAGUUUACAAUGUAAACGCGAAAAUGCGACAGCAGUGCAUAAAAAUCC --------------------.......................((((((((.....((((((...))))))((......)))))))..)))........... ( -12.80, z-score = -1.28, R) >droWil1.scaffold_180698 4687762 101 + 11422946 -CCUGUUCUUCACAUUGUUCAUUCCUUACUCUUACCACCAACUCGCCUGUCAGCCAGUUUACAAUGUAAACGCGAAAAUGCGACAGCAGUGCAUAAAAAUCC -.(((((....(((((((.....................((((.((......)).)))).)))))))...(((......)))..)))))............. ( -13.75, z-score = -0.31, R) >droVir3.scaffold_13049 338594 86 + 25233164 --------------CCCCAGUCCCCGGCCCCCAACCCUCCCU--GCUUGUCGGCCAGUUUACAAUGUAAACGCGAAAAUGCGACAGCAGUGCAUAAAAAUCC --------------...........((((..(((.(......--).)))..)))).((((((...))))))(((....(((....))).))).......... ( -19.10, z-score = -1.99, R) >droMoj3.scaffold_6680 23919121 89 - 24764193 -------------GCCCCUGCCUCCUGCUGCCGUCAUUCCACUGGCUUGUCAGCCAGUUUACAAUGUAAACGCGAAAAUGCGACAGCAGUGCAUAAAAAUCC -------------.....(((...((((((.(((..((((..(((((....)))))((((((...))))))).)))...))).)))))).)))......... ( -25.30, z-score = -2.79, R) >droGri2.scaffold_15110 1633951 101 + 24565398 ACCCCCCUCGCCUGCCCCCAGCCCCUUUCACCCUCCUGCCAUC-GCUUGUCAGCCAGUUUACAAUGUAAACGCGAAAAUGCGACAGCAGUGCAUAAAACUCC .........((((((.....((...((((......(((.((..-...)).)))...((((((...))))))..))))..))....)))).)).......... ( -18.50, z-score = -2.15, R) >consensus ______________CCCCCCCCCCCAUCCCCCCCCCUUCUAUC_GCCUGUCAGCCAGUUUACAAUGUAAACGCGAAAAUGCGACAGCAGUGCAUAAAAAUCC ............................................((.((((.....((((((...))))))((......))))))))............... (-12.63 = -12.38 + -0.25)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:54:59 2011