
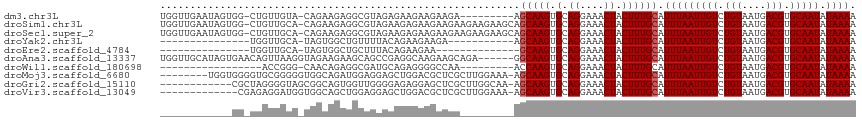
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 1,064,950 – 1,065,055 |
| Length | 105 |
| Max. P | 0.654723 |

| Location | 1,064,950 – 1,065,055 |
|---|---|
| Length | 105 |
| Sequences | 10 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 68.58 |
| Shannon entropy | 0.62674 |
| G+C content | 0.41831 |
| Mean single sequence MFE | -25.10 |
| Consensus MFE | -11.58 |
| Energy contribution | -11.68 |
| Covariance contribution | 0.10 |
| Combinations/Pair | 1.00 |
| Mean z-score | -0.98 |
| Structure conservation index | 0.46 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.29 |
| SVM RNA-class probability | 0.628394 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 1064950 105 + 24543557 UGGUUGAAUAGUGG-CUGUUGUA-CAGAAGAGGCGUAGAGAAGAAGAAGA---------AGCAAGUGCAGGAAACUACUUUGCAUUUAAUUGUCUGUAAUGACGUGCAAUAUAAAA ..............-.(((((((-(.........((((((..........---------.((....))((....)).))))))........(((......)))))))))))..... ( -22.00, z-score = -0.79, R) >droSim1.chr3L 691143 114 + 22553184 UGGUUGAAUAGUGG-CUGUUGCA-CAGAAGAGGCGUAGAAGAGAAGAAGAAGAAGAAGCAGCAAGUGCAGGAAACUACUUUGCAUUUAAUUGUCUGUAAUGACGUGCAAUAUAAAA ..............-.(((((((-(.......(((.((...................(((.....)))((....)).)).)))........(((......)))))))))))..... ( -21.90, z-score = 0.21, R) >droSec1.super_2 1068225 114 + 7591821 UGGUUGAAUAGUGG-CUGUUGCA-CAGAAGAGGCGUAGAAGAGAAGAAGAAGAAGAAGCAGCAAGUGCAGGAAACUACUUUGCAUUUAAUUGUCUGUAAUGACGUGCAAUAUAAAA ..............-.(((((((-(.......(((.((...................(((.....)))((....)).)).)))........(((......)))))))))))..... ( -21.90, z-score = 0.21, R) >droYak2.chr3L 1034307 89 + 24197627 ---------------UGGUUGCA-UAGUGGCUGUUUUACAGAAGAAGA-----------AGCAAGUGCAGGAAACUACUUUGCAUUUAAUUGUCUGUAAUGACGUGCAAUAUAAAA ---------------..((((((-(.......((((((((((....((-----------((((((.(.((....)).)))))).))).....))))))).))))))))))...... ( -21.51, z-score = -1.09, R) >droEre2.scaffold_4784 1062099 86 + 25762168 ---------------UGGUUGCA-UAGUGGCUGCUUUACAGAAGAA--------------GCAAGUGCAGGAAACUACUUUGCAUUUAAUUGUCUGUAAUGACGUGCAAUAUAAAA ---------------..((((((-((((....)))(((((((..((--------------..(((((((((.......)))))))))..)).)))))))....)))))))...... ( -21.90, z-score = -1.01, R) >droAna3.scaffold_13337 16441257 110 - 23293914 UGGUUGCAUAGUGAACAGUUAAGGUAGAAGAAGCAGCCGAGGCAAGAAGCAGA------GGCAAGUGCAGGAAACUACUUUGCAUUUAAUUGUCUGUAAUGACGUGCAAUAUAAAA ..(((((((.......(((((((...((((..((((((...((.....))...------)))...)))((....)).))))...)))))))(((......))))))))))...... ( -29.30, z-score = -1.97, R) >droWil1.scaffold_180698 6747063 89 - 11422946 -----------------ACCGGG-CAACAGAGGCGAUGCAGAGGGGCCAA---------ACCAAGUGCAGGAAACUACUUUGCAUUUAAUUGUCUGUAAUGACGUGCAAUAUAAAA -----------------.((.(.-...)...)).(((((((((((.....---------.))......((....)).)))))))))..(((((.(((....))).)))))...... ( -22.50, z-score = -0.69, R) >droMoj3.scaffold_6680 23908676 107 - 24764193 --------UGGUGGGGUGCGGGGGUGGCAGAUGGAGGAGCUGGACGCUCGCUUGGAAA-AGCAAGUGCAGGAAACUACUUUGCAUUUAAUUGUCUGUAAUGACGUGCAAUAUAAAA --------..(((..(..((..(...(((((..(..((((.....))))((((....)-)))(((((((((.......)))))))))..)..)))))..)..))..)..))).... ( -34.50, z-score = -3.07, R) >droGri2.scaffold_15110 1623462 103 + 24565398 ------------CGCUAGGGGUAGCGGCAGUGGUUGGGGAGAGGAGCUCGCUUGGCAA-AGCAAGUGCAGGAAACUACUUUGCAUUUAAUUGUCUGUAAUGACGUGCAAUAUAAAA ------------(((((....)))))(((((.((((.(((.((......((((....)-)))(((((((((.......)))))))))..)).))).)))).)).)))......... ( -29.30, z-score = -0.98, R) >droVir3.scaffold_13049 326903 102 + 25233164 -------------CGAGAGGAUGGUGGCAGCUGGAGGAGCUGGACGCUCGCUUGGAAA-AGCAAGUGCAGGAAACUACUUUGCAUUUAAUUGUCUGUAAUGACGUGCAAUAUAAAA -------------.........((((((((((.....)))))(.((((.((((....)-))).)))))......)))))((((((......(((......)))))))))....... ( -26.20, z-score = -0.61, R) >consensus ____________GG_CUGUUGCA_CAGAAGAGGCGUAGGAGAGAAGAAGA_________AGCAAGUGCAGGAAACUACUUUGCAUUUAAUUGUCUGUAAUGACGUGCAAUAUAAAA ............................................................(((((.(.((....)).)))))).(((((((((.(((....))).))))).)))). (-11.58 = -11.68 + 0.10)
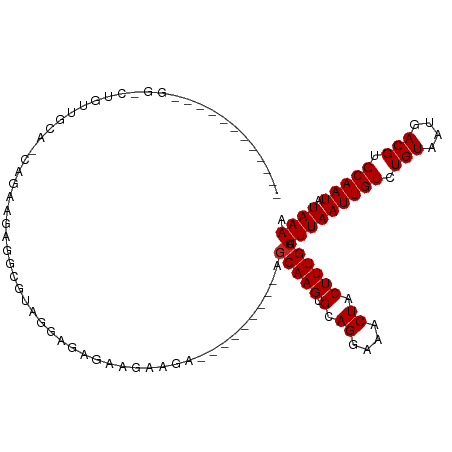

| Location | 1,064,950 – 1,065,055 |
|---|---|
| Length | 105 |
| Sequences | 10 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 68.58 |
| Shannon entropy | 0.62674 |
| G+C content | 0.41831 |
| Mean single sequence MFE | -16.22 |
| Consensus MFE | -7.53 |
| Energy contribution | -7.63 |
| Covariance contribution | 0.10 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.01 |
| Structure conservation index | 0.46 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.35 |
| SVM RNA-class probability | 0.654723 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 1064950 105 - 24543557 UUUUAUAUUGCACGUCAUUACAGACAAUUAAAUGCAAAGUAGUUUCCUGCACUUGCU---------UCUUCUUCUUCUCUACGCCUCUUCUG-UACAACAG-CCACUAUUCAACCA .........((..(((......)))........((((.((((....))))..)))).---------................)).....(((-.....)))-.............. ( -10.90, z-score = -0.32, R) >droSim1.chr3L 691143 114 - 22553184 UUUUAUAUUGCACGUCAUUACAGACAAUUAAAUGCAAAGUAGUUUCCUGCACUUGCUGCUUCUUCUUCUUCUUCUCUUCUACGCCUCUUCUG-UGCAACAG-CCACUAUUCAACCA .......(((((((((......)))........((((.((((....))))..))))...................................)-)))))...-.............. ( -16.40, z-score = -1.32, R) >droSec1.super_2 1068225 114 - 7591821 UUUUAUAUUGCACGUCAUUACAGACAAUUAAAUGCAAAGUAGUUUCCUGCACUUGCUGCUUCUUCUUCUUCUUCUCUUCUACGCCUCUUCUG-UGCAACAG-CCACUAUUCAACCA .......(((((((((......)))........((((.((((....))))..))))...................................)-)))))...-.............. ( -16.40, z-score = -1.32, R) >droYak2.chr3L 1034307 89 - 24197627 UUUUAUAUUGCACGUCAUUACAGACAAUUAAAUGCAAAGUAGUUUCCUGCACUUGCU-----------UCUUCUUCUGUAAAACAGCCACUA-UGCAACCA--------------- .......(((((.((..(((((((.........((((.((((....))))..)))).-----------......)))))))....)).....-)))))...--------------- ( -13.59, z-score = -0.79, R) >droEre2.scaffold_4784 1062099 86 - 25762168 UUUUAUAUUGCACGUCAUUACAGACAAUUAAAUGCAAAGUAGUUUCCUGCACUUGC--------------UUCUUCUGUAAAGCAGCCACUA-UGCAACCA--------------- .......((((..(((......))).............(((((...((((..((((--------------.......)))).))))..))))-)))))...--------------- ( -16.10, z-score = -1.06, R) >droAna3.scaffold_13337 16441257 110 + 23293914 UUUUAUAUUGCACGUCAUUACAGACAAUUAAAUGCAAAGUAGUUUCCUGCACUUGCC------UCUGCUUCUUGCCUCGGCUGCUUCUUCUACCUUAACUGUUCACUAUGCAACCA .......(((((.(((......))).............((((......(((...(((------...((.....))...)))))).....))))...............)))))... ( -18.40, z-score = -0.88, R) >droWil1.scaffold_180698 6747063 89 + 11422946 UUUUAUAUUGCACGUCAUUACAGACAAUUAAAUGCAAAGUAGUUUCCUGCACUUGGU---------UUGGCCCCUCUGCAUCGCCUCUGUUG-CCCGGU----------------- ............((.....(((((.......(((((.((..(((.((.......)).---------..)))..)).)))))....)))))..-..))..----------------- ( -14.10, z-score = 1.31, R) >droMoj3.scaffold_6680 23908676 107 + 24764193 UUUUAUAUUGCACGUCAUUACAGACAAUUAAAUGCAAAGUAGUUUCCUGCACUUGCU-UUUCCAAGCGAGCGUCCAGCUCCUCCAUCUGCCACCCCCGCACCCCACCA-------- ........(((..(((......)))........(((..((((....))))....(((-(....))))((((.....)))).......))).......)))........-------- ( -18.60, z-score = -2.35, R) >droGri2.scaffold_15110 1623462 103 - 24565398 UUUUAUAUUGCACGUCAUUACAGACAAUUAAAUGCAAAGUAGUUUCCUGCACUUGCU-UUGCCAAGCGAGCUCCUCUCCCCAACCACUGCCGCUACCCCUAGCG------------ .........((..(((......)))........((((((((((.......)).))))-))))..((((.((.................)))))).......)).------------ ( -19.33, z-score = -1.90, R) >droVir3.scaffold_13049 326903 102 - 25233164 UUUUAUAUUGCACGUCAUUACAGACAAUUAAAUGCAAAGUAGUUUCCUGCACUUGCU-UUUCCAAGCGAGCGUCCAGCUCCUCCAGCUGCCACCAUCCUCUCG------------- ..((((.(((((.(((......))).......))))).))))......((.((((((-(....))))))).)).(((((.....)))))..............------------- ( -18.40, z-score = -1.52, R) >consensus UUUUAUAUUGCACGUCAUUACAGACAAUUAAAUGCAAAGUAGUUUCCUGCACUUGCU_________UCUGCUUCUCUCCAACGCCUCUGCUA_UGCAACAG_CC____________ .............(((......)))........((((.((((....))))..))))............................................................ ( -7.53 = -7.63 + 0.10)



Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:54:58 2011