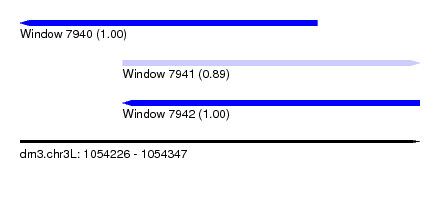
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 1,054,226 – 1,054,347 |
| Length | 121 |
| Max. P | 0.998896 |

| Location | 1,054,226 – 1,054,316 |
|---|---|
| Length | 90 |
| Sequences | 4 |
| Columns | 98 |
| Reading direction | reverse |
| Mean pairwise identity | 47.43 |
| Shannon entropy | 0.85414 |
| G+C content | 0.39872 |
| Mean single sequence MFE | -22.58 |
| Consensus MFE | -5.59 |
| Energy contribution | -5.02 |
| Covariance contribution | -0.56 |
| Combinations/Pair | 1.50 |
| Mean z-score | -3.12 |
| Structure conservation index | 0.25 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.49 |
| SVM RNA-class probability | 0.998790 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
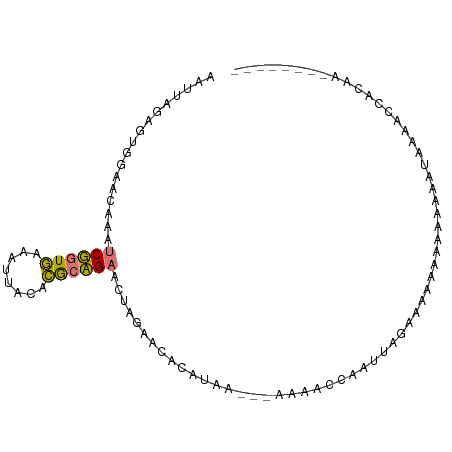
>dm3.chr3L 1054226 90 - 24543557 CACCGGUUUGGUCGAAAGCUCUAAAGCUACAUGCAGAGCUAGACCACUUGUUGCAAUAUCAGCAAGAAUUAAAGACCCAUAAGCUCGAGA-------- ....(((((((((...((((((...((.....)))))))).)))).(((((((......)))))))......))))).............-------- ( -26.90, z-score = -3.14, R) >droWil1.scaffold_180943 43144 89 + 149748 CAAUAAGUUGGAAUACCUCGGUGAGAUUACACGCCGAGGUAGAAUUCAUAA---AAAGCCAAUUGGGCGAAAAGAAAAGUAAAACCACAAAA------ ......((.((..((((((((((........))))))))))..........---...(((.....)))................))))....------ ( -24.10, z-score = -4.44, R) >dp4.Unknown_group_206 63046 90 + 92907 AUUUGCCGUGGAGCACAUCUGUGAAAUUACACACAGAACUCGAUCACAUCAAAUUGUUGCUAGCAGAAAAGAAAAUGAAAGAGCCAAUGC-------- ....((..(((......((((((........)))))).(((..(((..........(((....))).........)))..))))))..))-------- ( -15.11, z-score = 0.42, R) >droAna3.scaffold_10225 15752 95 + 63334 AAUUAGAGUGGAACAACUCGGUGAAAAUACACGCCGAAGUUGAACACAUAA---AAAACUUAUUAACGCAAAGAAGUAAUAAAACCACAGCCACUCAC .....((((((..((((((((((........))))).))))).........---.....((((((.(........)))))))........)))))).. ( -24.20, z-score = -5.31, R) >consensus AAUUAGAGUGGAACAAAUCGGUGAAAUUACACGCAGAACUAGAACACAUAA___AAAACCAAUUAGAAAAAAAAAAAAAUAAAACCACAA________ .................((((((........))))))............................................................. ( -5.59 = -5.02 + -0.56)

| Location | 1,054,257 – 1,054,347 |
|---|---|
| Length | 90 |
| Sequences | 4 |
| Columns | 94 |
| Reading direction | forward |
| Mean pairwise identity | 52.85 |
| Shannon entropy | 0.76484 |
| G+C content | 0.40011 |
| Mean single sequence MFE | -22.22 |
| Consensus MFE | -5.69 |
| Energy contribution | -7.38 |
| Covariance contribution | 1.69 |
| Combinations/Pair | 1.68 |
| Mean z-score | -2.04 |
| Structure conservation index | 0.26 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.11 |
| SVM RNA-class probability | 0.893300 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 1054257 90 + 24543557 ----AUAUUGCAACAAGUGGUCUAGCUCUGCAUGUAGCUUUAGAGCUUUCGACCAAACCGGUGGCGUUUGCUGACCGUAUAUUAGCCACCUUAC ----.............(((((.((((((((.....))...))))))...)))))....((((((...(((.....))).....)))))).... ( -28.40, z-score = -2.41, R) >droWil1.scaffold_180943 43174 87 - 149748 ----UUGGCUUUUUAUGAAUUCUACCUCGGCGUGUAAUCUCACCGAGGUAUUCCA---ACUUAUUGUAUUCGGGCUGUGUAAUAGCCUCAAAAU ----(((.........((((..((((((((.(.(....).).)))))))).....---((.....))))))(((((((...))))))))))... ( -21.30, z-score = -1.95, R) >dp4.Unknown_group_206 63077 87 - 92907 ----CAACAAUUUGAUGUGAUCGAGUUCUGUGUGUAAUUUCACAGAUGUGCUCCA---CGGCAAAUUAUCGUUGCUAUUUAGCAGUUUGAGUAG ----((((((((((.((((...((((((((((........))))))...))))))---)).))))))...))))((((((((....)))))))) ( -21.90, z-score = -1.26, R) >droAna3.scaffold_10225 15784 91 - 63334 UUAAUAAGUUUUUUAUGUGUUCAACUUCGGCGUGUAUUUUCACCGAGUUGUUCCA---CUCUAAUUUACUUCCGCUAUUUAGUAGCUUCAGUAA ................(((..(((((.(((.(........).))))))))...))---)......(((((...(((((...)))))...))))) ( -17.30, z-score = -2.56, R) >consensus ____UAAGUUUUUUAUGUGUUCUACCUCGGCGUGUAAUUUCACAGAGGUGUUCCA___CGGUAAUGUAUUCUGGCUAUUUAGUAGCCUCAGUAA .......................(((((((.(........).)))))))................(((((.((((((.....)))))).))))) ( -5.69 = -7.38 + 1.69)

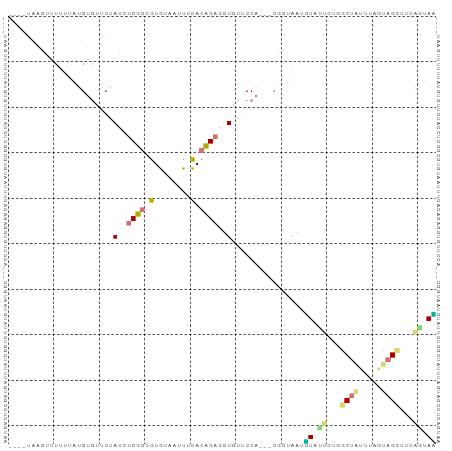
| Location | 1,054,257 – 1,054,347 |
|---|---|
| Length | 90 |
| Sequences | 4 |
| Columns | 94 |
| Reading direction | reverse |
| Mean pairwise identity | 52.85 |
| Shannon entropy | 0.76484 |
| G+C content | 0.40011 |
| Mean single sequence MFE | -24.58 |
| Consensus MFE | -5.79 |
| Energy contribution | -6.10 |
| Covariance contribution | 0.31 |
| Combinations/Pair | 1.55 |
| Mean z-score | -3.57 |
| Structure conservation index | 0.24 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.54 |
| SVM RNA-class probability | 0.998896 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
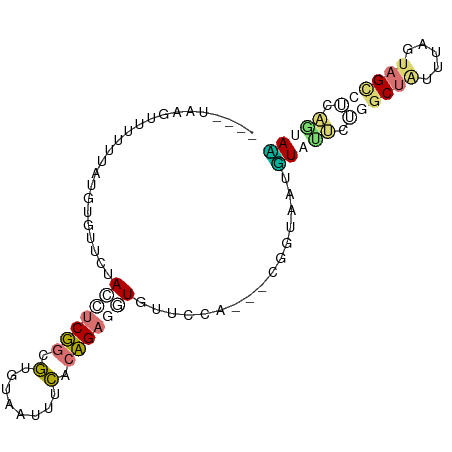
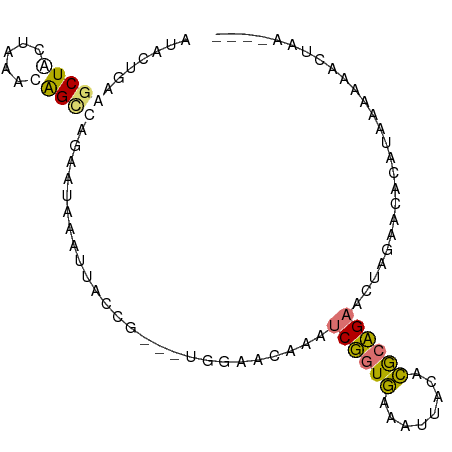
>dm3.chr3L 1054257 90 - 24543557 GUAAGGUGGCUAAUAUACGGUCAGCAAACGCCACCGGUUUGGUCGAAAGCUCUAAAGCUACAUGCAGAGCUAGACCACUUGUUGCAAUAU---- ((((((((((...................))))))....(((((...((((((...((.....)))))))).)))))....)))).....---- ( -28.51, z-score = -2.20, R) >droWil1.scaffold_180943 43174 87 + 149748 AUUUUGAGGCUAUUACACAGCCCGAAUACAAUAAGU---UGGAAUACCUCGGUGAGAUUACACGCCGAGGUAGAAUUCAUAAAAAGCCAA---- ..((((.((((.......))))))))..........---((((.((((((((((........))))))))))...))))...........---- ( -25.20, z-score = -4.06, R) >dp4.Unknown_group_206 63077 87 + 92907 CUACUCAAACUGCUAAAUAGCAACGAUAAUUUGCCG---UGGAGCACAUCUGUGAAAUUACACACAGAACUCGAUCACAUCAAAUUGUUG---- ..........((((....)))).((((((((((..(---(((......((((((........))))))......))))..))))))))))---- ( -21.00, z-score = -3.23, R) >droAna3.scaffold_10225 15784 91 + 63334 UUACUGAAGCUACUAAAUAGCGGAAGUAAAUUAGAG---UGGAACAACUCGGUGAAAAUACACGCCGAAGUUGAACACAUAAAAAACUUAUUAA (((((...((((.....))))...)))))......(---((...((((((((((........))))).)))))..)))................ ( -23.60, z-score = -4.80, R) >consensus AUACUGAAGCUACUAAACAGCCAGAAUAAAUUACCG___UGGAACAAAUCGGUGAAAUUACACGCAGAACUAGAACACAUAAAAAACUAA____ .......(((((.....)))))..........................((((((........)))))).......................... ( -5.79 = -6.10 + 0.31)

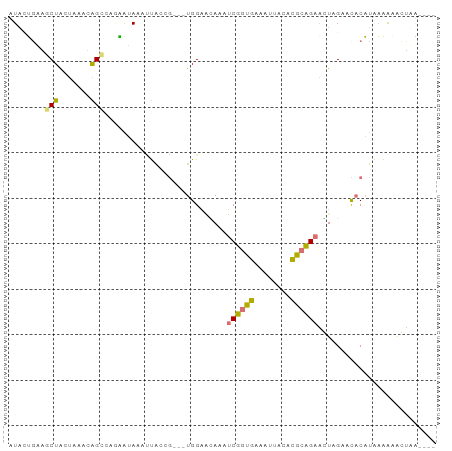
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:54:56 2011