| Sequence ID | dm3.chr3L |
|---|---|
| Location | 1,019,512 – 1,019,607 |
| Length | 95 |
| Max. P | 0.619506 |

| Location | 1,019,512 – 1,019,607 |
|---|---|
| Length | 95 |
| Sequences | 7 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 72.50 |
| Shannon entropy | 0.51066 |
| G+C content | 0.36609 |
| Mean single sequence MFE | -25.40 |
| Consensus MFE | -9.66 |
| Energy contribution | -10.26 |
| Covariance contribution | 0.60 |
| Combinations/Pair | 1.43 |
| Mean z-score | -1.92 |
| Structure conservation index | 0.38 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.26 |
| SVM RNA-class probability | 0.619506 |
| Prediction | RNA |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
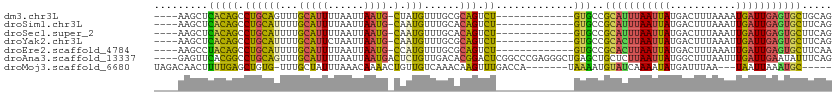
>dm3.chr3L 1019512 95 + 24543557 ----AAGCUCACAGCCUGCAGUUUGCAUUUUAAUUAAUG-CUAUGUUUGCGCAGUCU-------------GUGCCGCAUUUAAUUAUGACUUUAAAAUGAUUGAGUGCUGCAG ----..((.(((((.((((.((..((((..((.....))-..))))..)))))).))-------------)))..((((((((((((.........)))))))))))).)).. ( -29.60, z-score = -2.45, R) >droSim1.chr3L 652414 95 + 22553184 ----AAGCUCACAGCCUGCAUUUUGCAUUUUAAUUAAUG-CAAUGUUUGCACAGUCU-------------GUGCCGCAUUUAAUUAUGACUUUAAAUUGAUUGAGUGCUUCAG ----.....((((((.((((..(((((((......))))-)))....))))..).))-------------)))..(((((((((((...........)))))))))))..... ( -26.20, z-score = -2.26, R) >droSec1.super_2 1031004 95 + 7591821 ----AAGCUCACAGCCUGCAUUUUGCAUUUUAAUUAAUG-CAAUGUUUGCACAGUCU-------------GUGCCGCAUUUAAUUAUGACUUUAAAUUGAUUGAGUGCUUCAG ----.....((((((.((((..(((((((......))))-)))....))))..).))-------------)))..(((((((((((...........)))))))))))..... ( -26.20, z-score = -2.26, R) >droYak2.chr3L 996009 95 + 24197627 ----AAGCUCACAGCCUGCAUUUUGCAUUCUAAUUAAUG-CAAUGUUUGCGCAGUCU-------------GUGCCGCACUUAAUUAUGACUUUAAAUUGAUUGAGUGCUUCAG ----.....(((((.((((...(((((((......))))-))).(....))))).))-------------)))..(((((((((((...........)))))))))))..... ( -29.70, z-score = -3.22, R) >droEre2.scaffold_4784 1021819 95 + 25762168 ----AAGCCUACAGCCUGCAUUUUGCAUUUUAAUUAAUG-CCAUGUUUGCGCAGUCU-------------GUGCCGCACUUAAUUAUGACUUUAAAUUGAUUGAGUGCUUCAA ----.....(((((.((((.....(((((......))))-)((....)).)))).))-------------)))..(((((((((((...........)))))))))))..... ( -26.50, z-score = -2.82, R) >droAna3.scaffold_13337 16402498 109 - 23293914 ----GAGUUCACGGCCUGCAGUUUGCAUUUUAAUUAAUGACUCUGUUGACACGGACUCGGCCCGAGGGCUGAGCUGCUCUUAAUUAUGGCUUUAAUUUGAUUGAAUAUUUCAG ----(((((...((((.((((....((((......))))...)))).....(((.(((((((....))))))))))...........))))..)))))............... ( -27.20, z-score = -0.86, R) >droMoj3.scaffold_6680 23863513 97 - 24764193 UAGACAACUUUUGAGCUGUG-UUUGCUAUUUAAACAAAACUGUUGUCAAACAAGUUUGACCA-------UAAAAUGUAUCAAAAUAUGAUUUAA---UAAUUAAAUGC----- ((((((..((((((....((-((((.....))))))....(((.((((((....)))))).)-------)).......))))))..)).)))).---...........----- ( -12.40, z-score = 0.45, R) >consensus ____AAGCUCACAGCCUGCAUUUUGCAUUUUAAUUAAUG_CAAUGUUUGCACAGUCU_____________GUGCCGCAUUUAAUUAUGACUUUAAAUUGAUUGAGUGCUUCAG .............((..((((....((((......))))...))))..)).........................(((((((((((...........)))))))))))..... ( -9.66 = -10.26 + 0.60)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:54:54 2011