
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 975,841 – 975,943 |
| Length | 102 |
| Max. P | 0.999793 |

| Location | 975,841 – 975,943 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 72.58 |
| Shannon entropy | 0.50348 |
| G+C content | 0.45404 |
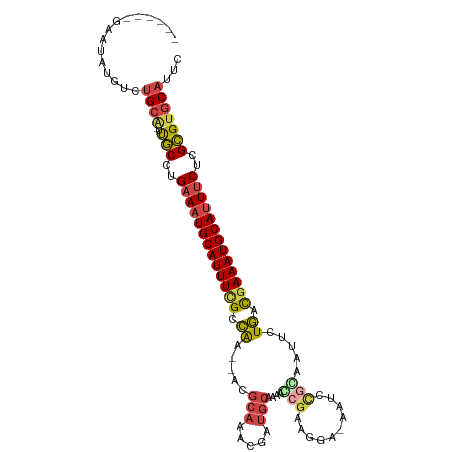
| Mean single sequence MFE | -34.30 |
| Consensus MFE | -20.36 |
| Energy contribution | -21.03 |
| Covariance contribution | 0.67 |
| Combinations/Pair | 1.36 |
| Mean z-score | -3.29 |
| Structure conservation index | 0.59 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.98 |
| SVM RNA-class probability | 0.999526 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
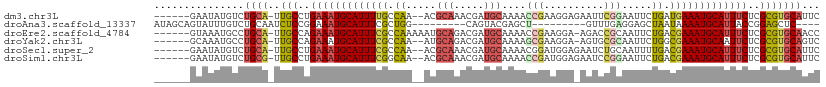
>dm3.chr3L 975841 102 + 24543557 ------GAAUAUGUCUGCA-UUGCCUGAAAUGCAUUUUGCCAA--ACGCAAACGAUGCAAAACCGAAGGAGAAUUCGGAAUUCUGAUGAAAUGCAUUUCUCGCGUGCAUUC ------.........((((-(.((..((((((((((((..((.--..(((.....)))....(((((......))))).....))..))))))))))))..)))))))... ( -32.80, z-score = -3.35, R) >droAna3.scaffold_13337 16351783 89 - 23293914 AUAGCAGUAUUUGUCUGCAAUCUCCGGAAAUGCAUUUCGCUGG---------CAGUACGAGCU---------GUUUGAGGAGCUAAUAAAAUGCAUUACCGGAGCUC---- ...((((.......))))...((((((.(((((((((.(((((---------((((....)))---------))).....))).....))))))))).))))))...---- ( -30.90, z-score = -2.92, R) >droEre2.scaffold_4784 976647 103 + 25762168 ------GUAAAUGCCUGCA-UUGCCAGAAAUGCAUUUCGCCAAAAAUGCAGACGAUGCAAAACCGAAGGA-AGACCGCAAUUCUGACGAAAUGCAUUUCUCGCGUGCAACC ------.........((((-(.((.((((((((((((((.((.((.(((...((.........))..((.-...))))).)).)).)))))))))))))).)))))))... ( -33.90, z-score = -4.04, R) >droYak2.chr3L 951263 101 + 24197627 ------GCAAAUGCCUGCA-UUGCCAGAAAUGCAUUUCGCCAA--AUGCAGACGAUGCAAAAGCGAAGGA-AGUGCGCAAUUCUGGCGAAAUGCAAUUCUCGCGUGCAGUC ------((....))(((((-(.((.((((.((((((((((((.--.((((.....))))...(((.....-....))).....)))))))))))).)))).)))))))).. ( -42.20, z-score = -3.88, R) >droSec1.super_2 987792 102 + 7591821 ------GAAUAUGUCUGCA-UUGCCUGAAAUGCAUUUCGCCAA--ACGCAAACGAUGCAAAACGGAUGGAGAAUCUGCAAUUUUGACGAAAUGCAUUUCUCGCGUGCAUUC ------.........((((-(.((..(((((((((((((.(((--(.(((.....)))....(((((.....)))))....)))).)))))))))))))..)))))))... ( -35.20, z-score = -3.69, R) >droSim1.chr3L 607612 102 + 22553184 ------GAAUAUGUCUGCG-UUGCCUGAAAUGCAUUUCGGCAA--ACGCAAACGAUGCAAAACCGAUGGAGAAUCCGGAAUUCUGACGAAAUGCAUUUCUCGCGUGCAUUC ------....((((.((((-......(((((((((((((.((.--..(((.....)))....(((((.....)).))).....)).))))))))))))).)))).)))).. ( -30.80, z-score = -1.88, R) >consensus ______GAAUAUGUCUGCA_UUGCCUGAAAUGCAUUUCGCCAA__ACGCAAACGAUGCAAAACCGAAGGA_AAUCCGCAAUUCUGACGAAAUGCAUUUCUCGCGUGCAUUC ...............((((..(((..(((((((((((((.((.....(((.....)))....(((..........))).....)).)))))))))))))..)))))))... (-20.36 = -21.03 + 0.67)
| Location | 975,841 – 975,943 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 72.58 |
| Shannon entropy | 0.50348 |
| G+C content | 0.45404 |
| Mean single sequence MFE | -36.28 |
| Consensus MFE | -22.47 |
| Energy contribution | -24.45 |
| Covariance contribution | 1.98 |
| Combinations/Pair | 1.35 |
| Mean z-score | -3.68 |
| Structure conservation index | 0.62 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 4.41 |
| SVM RNA-class probability | 0.999793 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
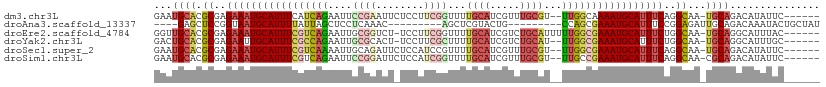
>dm3.chr3L 975841 102 - 24543557 GAAUGCACGCGAGAAAUGCAUUUCAUCAGAAUUCCGAAUUCUCCUUCGGUUUUGCAUCGUUUGCGU--UUGGCAAAAUGCAUUUCAGGCAA-UGCAGACAUAUUC------ (..((((.((..(((((((((((..(((((...(((((......)))))....(((.....))).)--))))..)))))))))))..))..-))))..)......------ ( -30.10, z-score = -2.16, R) >droAna3.scaffold_13337 16351783 89 + 23293914 ----GAGCUCCGGUAAUGCAUUUUAUUAGCUCCUCAAAC---------AGCUCGUACUG---------CCAGCGAAAUGCAUUUCCGGAGAUUGCAGACAAAUACUGCUAU ----...((((((.((((((((((...((((........---------)))).((....---------...)))))))))))).))))))...((((.......))))... ( -28.40, z-score = -3.34, R) >droEre2.scaffold_4784 976647 103 - 25762168 GGUUGCACGCGAGAAAUGCAUUUCGUCAGAAUUGCGGUCU-UCCUUCGGUUUUGCAUCGUCUGCAUUUUUGGCGAAAUGCAUUUCUGGCAA-UGCAGGCAUUUAC------ (.(((((.((.((((((((((((((((((((.(((((...-.....((((.....)))).))))).)))))))))))))))))))).))..-))))).)......------ ( -47.40, z-score = -6.49, R) >droYak2.chr3L 951263 101 - 24197627 GACUGCACGCGAGAAUUGCAUUUCGCCAGAAUUGCGCACU-UCCUUCGCUUUUGCAUCGUCUGCAU--UUGGCGAAAUGCAUUUCUGGCAA-UGCAGGCAUUUGC------ ..(((((.((.((((.((((((((((((((...(((....-.....)))...((((.....)))))--))))))))))))).)))).))..-)))))........------ ( -42.70, z-score = -4.14, R) >droSec1.super_2 987792 102 - 7591821 GAAUGCACGCGAGAAAUGCAUUUCGUCAAAAUUGCAGAUUCUCCAUCCGUUUUGCAUCGUUUGCGU--UUGGCGAAAUGCAUUUCAGGCAA-UGCAGACAUAUUC------ (..((((.((..((((((((((((((((((...((((((....((.......))....)))))).)--)))))))))))))))))..))..-))))..)......------ ( -37.00, z-score = -3.78, R) >droSim1.chr3L 607612 102 - 22553184 GAAUGCACGCGAGAAAUGCAUUUCGUCAGAAUUCCGGAUUCUCCAUCGGUUUUGCAUCGUUUGCGU--UUGCCGAAAUGCAUUUCAGGCAA-CGCAGACAUAUUC------ (..(((......((((((((((((...........((.....))...(((..((((.....)))).--..))))))))))))))).(....-))))..)......------ ( -32.10, z-score = -2.14, R) >consensus GAAUGCACGCGAGAAAUGCAUUUCGUCAGAAUUGCGGACU_UCCUUCGGUUUUGCAUCGUUUGCGU__UUGGCGAAAUGCAUUUCAGGCAA_UGCAGACAUAUUC______ ...((((.((..(((((((((((((((((....((((........))))...((((.....))))...)))))))))))))))))..))...))))............... (-22.47 = -24.45 + 1.98)
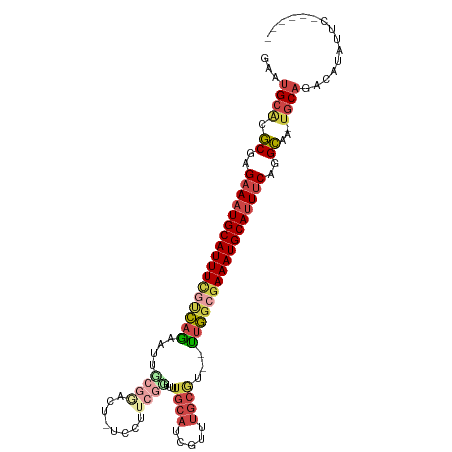
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:54:47 2011