| Sequence ID | dm3.chr3L |
|---|---|
| Location | 961,696 – 961,830 |
| Length | 134 |
| Max. P | 0.885608 |

| Location | 961,696 – 961,794 |
|---|---|
| Length | 98 |
| Sequences | 6 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 56.51 |
| Shannon entropy | 0.77771 |
| G+C content | 0.33169 |
| Mean single sequence MFE | -19.07 |
| Consensus MFE | -7.93 |
| Energy contribution | -6.82 |
| Covariance contribution | -1.11 |
| Combinations/Pair | 1.64 |
| Mean z-score | -1.01 |
| Structure conservation index | 0.42 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.07 |
| SVM RNA-class probability | 0.885608 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
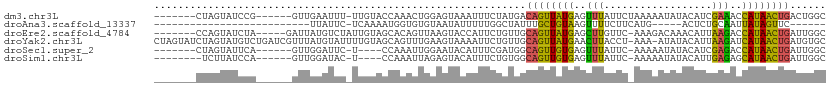
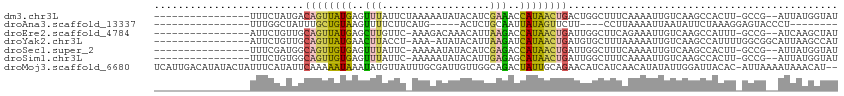
>dm3.chr3L 961696 98 + 24543557 -------CUAGUAUCCG------GUUGAAUUU-UUGUACCAAACUGGAGUAAAUUUCUAUGACAGUUAUGAGUUUAUUCUAAAAAUAUACAUCGAAACCAUAACUGACUGGC -------(((((.((((------(((......-........)))))))..............((((((((.((((...................))))))))))))))))). ( -21.55, z-score = -2.16, R) >droAna3.scaffold_13337 16336456 74 - 23293914 --------------------------UUAUUC-UCAAAAUGGUGUGUAAUAUUUUUGGCUAUUUGCUGUAAGUUUUCUUCAUG-----ACUCUGCAAUUAUAGUUC------ --------------------------......-.(((((..((.....))..)))))(((((((((....((((........)-----)))..))))..)))))..------ ( -8.40, z-score = 0.29, R) >droEre2.scaffold_4784 962874 99 + 25762168 -------CCAGUAUCUA-----GAUUAUGUCUAUUGUAGCACAGUUAAGUACCAUUCUGUUGCAGUUAUGAGCUUGUUC-AAAGACAAACAUUAAGACCAUAACUGAUUGGC -------(((((...((-----(.(((((((((((((((((.(((........))).))))))))).....(.(((((.-...))))).)....))).))))))))))))). ( -18.60, z-score = 0.09, R) >droYak2.chr3L 937187 110 + 24197627 CUAGUAUCUAGUAUGUCUGAUCGUUUAUGUAUUUUGUAGCAGUUUGAAGUAAAAUUCUGUUGCAGUUAUGAACUUACCU-AAA-AUAUACAUUAAGAUCAUAACUGAUGUGC ..........(((((((..................(((((((..(........)..)))))))((((((((.((((...-...-........)))).))))))))))))))) ( -22.64, z-score = -1.22, R) >droSec1.super_2 973408 93 + 7591821 -------CUAGUAUUCA------GUUGGAUUC-U----CCAAAUUGGAAUACAUUUCGAUGGCAGUUGUGAGUUUAUUC-AAAAAUAUACAUCGAGACCAUAACUGAUUGGC -------...((((((.------.((((....-.----))))....)))))).((((((((.......((((....)))-)........))))))))(((........))). ( -23.36, z-score = -2.21, R) >droSim1.chr3L 593673 92 + 22553184 --------UCUUAUCCA------GUUGGAUAC-U----CCAAAUUAGAGUACAUUUCUGUGGCAGUUGUGAGUUUAUUC-AAAAAUAUACAUUGAGAGCAUAACUGAUUGGC --------...(((((.------...))))).-.----((((..(((((.....)))))...((((((((..((((...-............))))..)))))))).)))). ( -19.86, z-score = -0.87, R) >consensus _______CUAGUAUCCA______GUUGUAUUC_UUGUACCAAAUUGGAGUACAUUUCUGUGGCAGUUAUGAGUUUAUUC_AAAAAUAUACAUUGAGACCAUAACUGAUUGGC ..............................................................((((((((..(((..................)))..))))))))...... ( -7.93 = -6.82 + -1.11)
| Location | 961,735 – 961,830 |
|---|---|
| Length | 95 |
| Sequences | 7 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 54.55 |
| Shannon entropy | 0.82974 |
| G+C content | 0.32789 |
| Mean single sequence MFE | -18.83 |
| Consensus MFE | -5.73 |
| Energy contribution | -5.10 |
| Covariance contribution | -0.63 |
| Combinations/Pair | 1.73 |
| Mean z-score | -1.22 |
| Structure conservation index | 0.30 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.32 |
| SVM RNA-class probability | 0.642520 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
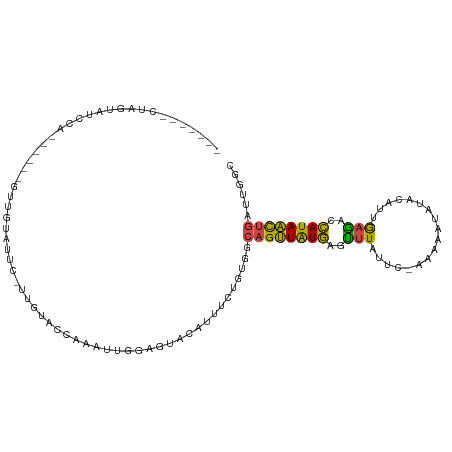
>dm3.chr3L 961735 95 + 24543557 ----------------UUUCUAUGACAGUUAUGAGUUUAUUCUAAAAAUAUACAUCGAAACCAUAACUGACUGGCUUUCAAAAUUGUCAAGCCACUU-GCCG--AUUAUGGUAU ----------------...((((((((((((((.((((...................))))))))))))..((((((.((....))..))))))...-....--.))))))... ( -19.41, z-score = -2.22, R) >droAna3.scaffold_13337 16336482 81 - 23293914 ----------------UUUGGCUAUUUGCUGUAAGUUUUCUUCAUG-----ACUCUGCAAUUAUAGUUCUU----CCUUAAAAUUAAUAUUCUAAAGGAGUACCCU-------- ----------------...((((((((((....((((........)-----)))..))))..)))))).((----((((...............))))))......-------- ( -10.76, z-score = -0.05, R) >droEre2.scaffold_4784 962915 94 + 25762168 ----------------AUUCUGUUGCAGUUAUGAGCUUGUUC-AAAGACAAACAUUAAGACCAUAACUGAUUGGCUUCAGAAAUUGUCAAGCCAUUU-GCCG--AUCAAGCUAU ----------------........(((((((((..((((...-............))))..)))))))(((((((......................-))))--)))..))... ( -19.71, z-score = -0.66, R) >droYak2.chr3L 937240 96 + 24197627 ----------------AUUCUGUUGCAGUUAUGAACUUACCU-AAA-AUAUACAUUAAGAUCAUAACUGAUGUGCUUUAAAAAUUGUCAAGCCAUUUUGGCGGCAUUAAGCCAU ----------------.........(((((((((.((((...-...-........)))).)))))))))(((.((((.......((((..(((.....)))))))..))))))) ( -22.34, z-score = -2.22, R) >droSec1.super_2 973443 94 + 7591821 ----------------UUUCGAUGGCAGUUGUGAGUUUAUUC-AAAAAUAUACAUCGAGACCAUAACUGAUUGGCUUUCAAAAUUGUCAAGCCACUU-GCCG--AUUAUGGUAU ----------------.(((((((.......((((....)))-)........)))))))((((((((.(..((((((.((....))..))))))...-.).)--.))))))).. ( -22.36, z-score = -1.72, R) >droSim1.chr3L 593707 94 + 22553184 ----------------UUUCUGUGGCAGUUGUGAGUUUAUUC-AAAAAUAUACAUUGAGAGCAUAACUGAUUGGCUUUCAAAAUUGUCAAGCCACUU-GCCG--AUUAUGGUAU ----------------.....((((((((((((..((((...-............))))..)))))))..(((((..........)))))))))).(-(((.--.....)))). ( -23.16, z-score = -1.58, R) >droMoj3.scaffold_6680 21325619 111 + 24764193 UCAUUGACAUAUACUAUUUCAUAUUCAAAAAUAAAUAUGUUAUUUGCGAUUGUUGGCAGACUAUUGCAGAACAUCAUCAACAUAUAUUGGAUUACAC-AUUAAAAUAAACAU-- ....((((((((..(((((.........))))).))))))))((((((((.(((....))).))))))))...........................-..............-- ( -14.10, z-score = -0.08, R) >consensus ________________UUUCUGUGGCAGUUAUGAGUUUAUUC_AAAAAUAUACAUUGAGACCAUAACUGAUUGGCUUUAAAAAUUGUCAAGCCACUU_GCCG__AUUAAGGUAU .........................((((((((..(((..................)))..))))))))............................................. ( -5.73 = -5.10 + -0.63)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:54:45 2011