| Sequence ID | dm3.chr3L |
|---|---|
| Location | 953,904 – 953,995 |
| Length | 91 |
| Max. P | 0.930152 |

| Location | 953,904 – 953,995 |
|---|---|
| Length | 91 |
| Sequences | 7 |
| Columns | 100 |
| Reading direction | forward |
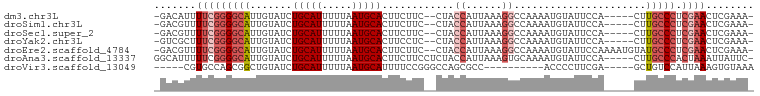
| Mean pairwise identity | 77.67 |
| Shannon entropy | 0.43446 |
| G+C content | 0.42314 |
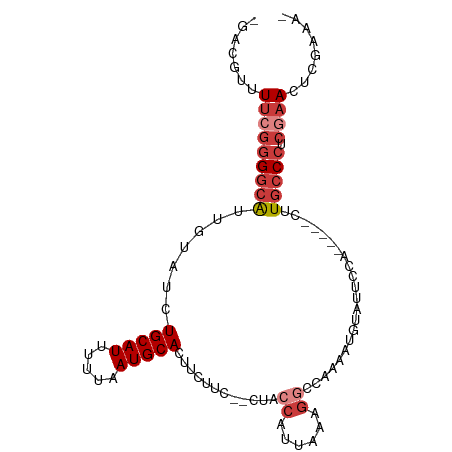
| Mean single sequence MFE | -23.45 |
| Consensus MFE | -13.24 |
| Energy contribution | -14.83 |
| Covariance contribution | 1.59 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.75 |
| Structure conservation index | 0.56 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.81 |
| SVM RNA-class probability | 0.825352 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
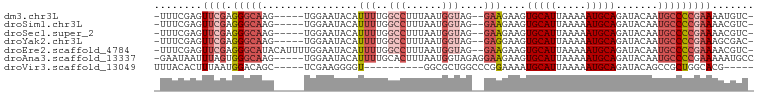
>dm3.chr3L 953904 91 + 24543557 -UUUCGAGUUCGAGGGCAAG-----UGGAAUACAUUUUGGCCUUUAAUGGUAG--GAAGAAGUGCAUUAAAAAUGCAGAUACAAUGCCCCGAAAAUGUC- -...((..((((.(((((..-----((......(((((..(((........))--)..)))))((((.....)))).....)).)))))))))..))..- ( -22.70, z-score = -1.65, R) >droSim1.chr3L 585608 91 + 22553184 -UUUCGAGUUCGAGGGCAAG-----UGGAAUACAUUUUGGCCUUUAAUGGUAG--GAAGAAGUGCAUUAAAAAUGCAGAUACAAUGCCCCGAAAACGUC- -...((..((((.(((((..-----((......(((((..(((........))--)..)))))((((.....)))).....)).)))))))))..))..- ( -24.30, z-score = -2.14, R) >droSec1.super_2 964500 91 + 7591821 -UUUCGAGUUCGAGGGCAAG-----UGGAAUACAUUUUGGCCUUUAAUGGUAG--GAAGAAGUGCAUUAAAAAUGCAGAUACAAUGCCCCGAAAACGUC- -...((..((((.(((((..-----((......(((((..(((........))--)..)))))((((.....)))).....)).)))))))))..))..- ( -24.30, z-score = -2.14, R) >droYak2.chr3L 927882 91 + 24197627 -UUUCGAGUUCGAGGGCAAG-----UGGAAUACAUUUUGGCCUUUAAUGGUAG--GAGGAAGUGCAUUAAAAAUGCAGAUACAAUGCCCCGAAAGCGAC- -..(((..((((.(((((..-----((......(((((..(((........))--)..)))))((((.....)))).....)).)))))))))..))).- ( -27.20, z-score = -2.90, R) >droEre2.scaffold_4784 955174 96 + 25762168 -UUUCGAGUUCGAGGGCAUACAUUUUGGAAUACAUUUUGGCCUUUAAUGGUAG--GAAGAAGUGCAUUAAAAAUGCAGAUACAAUGCCCCGAAAACGUC- -...((..((((.((((((.((((((..(((.((((((..(((........))--)..)))))).))).))))))........))))))))))..))..- ( -26.60, z-score = -2.64, R) >droAna3.scaffold_13337 16328733 94 - 23293914 -GAAUAAUUUAGUGGGCAAG-----UGGAAUACAUUUUGCACUUUAAUGGUAGAGGAAGAAGUGCAUUAAAAAUGCAGAUACAAUGCCCCGAAAAAUGCC -..........(.(((((.(-----((.....(((((((((((((.............))))))))....)))))....)))..)))))).......... ( -19.22, z-score = -1.01, R) >droVir3.scaffold_13049 15511097 80 + 25233164 UUUACACUUUAAUGGACAGC-----UCGAAGGGGU----------GGCGCUGGCCCGGAAAAUGCAUUAAAAAUGCAGAUACAGCCGCUGGCACG----- ..............(...((-----((....))))----------..)((..((..((....(((((.....))))).......))))..))...----- ( -19.80, z-score = 0.20, R) >consensus _UUUCGAGUUCGAGGGCAAG_____UGGAAUACAUUUUGGCCUUUAAUGGUAG__GAAGAAGUGCAUUAAAAAUGCAGAUACAAUGCCCCGAAAACGUC_ ........((((.(((((.....................(((......)))...........(((((.....))))).......)))))))))....... (-13.24 = -14.83 + 1.59)
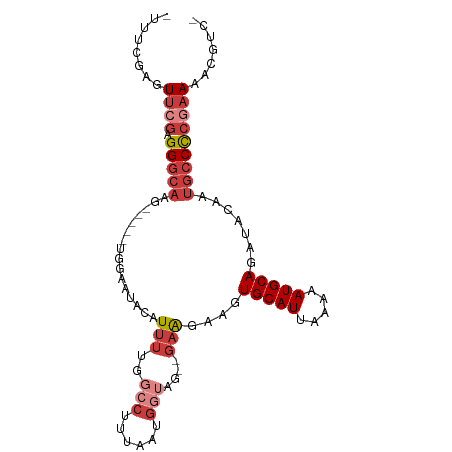
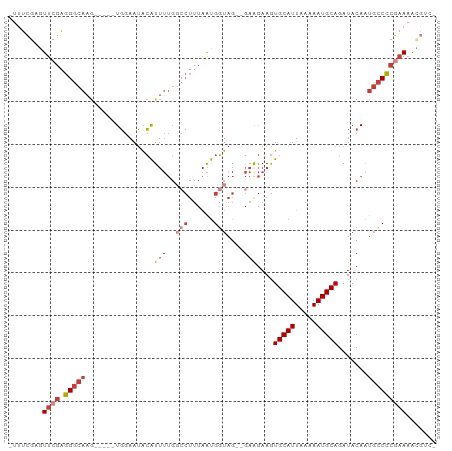
| Location | 953,904 – 953,995 |
|---|---|
| Length | 91 |
| Sequences | 7 |
| Columns | 100 |
| Reading direction | reverse |
| Mean pairwise identity | 77.67 |
| Shannon entropy | 0.43446 |
| G+C content | 0.42314 |
| Mean single sequence MFE | -21.11 |
| Consensus MFE | -10.52 |
| Energy contribution | -11.83 |
| Covariance contribution | 1.31 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.27 |
| Structure conservation index | 0.50 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.35 |
| SVM RNA-class probability | 0.930152 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 953904 91 - 24543557 -GACAUUUUCGGGGCAUUGUAUCUGCAUUUUUAAUGCACUUCUUC--CUACCAUUAAAGGCCAAAAUGUAUUCCA-----CUUGCCCUCGAACUCGAAA- -......(((((((((.......((((((((....((.(((....--.........))))).)))))))).....-----..))))).)))).......- ( -18.66, z-score = -1.89, R) >droSim1.chr3L 585608 91 - 22553184 -GACGUUUUCGGGGCAUUGUAUCUGCAUUUUUAAUGCACUUCUUC--CUACCAUUAAAGGCCAAAAUGUAUUCCA-----CUUGCCCUCGAACUCGAAA- -..((..(((((((((.......((((((((....((.(((....--.........))))).)))))))).....-----..))))).))))..))...- ( -21.66, z-score = -2.71, R) >droSec1.super_2 964500 91 - 7591821 -GACGUUUUCGGGGCAUUGUAUCUGCAUUUUUAAUGCACUUCUUC--CUACCAUUAAAGGCCAAAAUGUAUUCCA-----CUUGCCCUCGAACUCGAAA- -..((..(((((((((.......((((((((....((.(((....--.........))))).)))))))).....-----..))))).))))..))...- ( -21.66, z-score = -2.71, R) >droYak2.chr3L 927882 91 - 24197627 -GUCGCUUUCGGGGCAUUGUAUCUGCAUUUUUAAUGCACUUCCUC--CUACCAUUAAAGGCCAAAAUGUAUUCCA-----CUUGCCCUCGAACUCGAAA- -.(((..(((((((((.......((((((((....((.(((....--.........))))).)))))))).....-----..))))).))))..)))..- ( -22.16, z-score = -2.90, R) >droEre2.scaffold_4784 955174 96 - 25762168 -GACGUUUUCGGGGCAUUGUAUCUGCAUUUUUAAUGCACUUCUUC--CUACCAUUAAAGGCCAAAAUGUAUUCCAAAAUGUAUGCCCUCGAACUCGAAA- -..((..((((((((((.......(((((((.((((((......(--((........)))......))))))..))))))))))))).))))..))...- ( -25.21, z-score = -3.34, R) >droAna3.scaffold_13337 16328733 94 + 23293914 GGCAUUUUUCGGGGCAUUGUAUCUGCAUUUUUAAUGCACUUCUUCCUCUACCAUUAAAGUGCAAAAUGUAUUCCA-----CUUGCCCACUAAAUUAUUC- ...........(((((.......(((((((....(((((((...............)))))))))))))).....-----..)))))............- ( -20.20, z-score = -2.71, R) >droVir3.scaffold_13049 15511097 80 - 25233164 -----CGUGCCAGCGGCUGUAUCUGCAUUUUUAAUGCAUUUUCCGGGCCAGCGCC----------ACCCCUUCGA-----GCUGUCCAUUAAAGUGUAAA -----.......((((......))))........((((((((...((.((((...----------..........-----)))).))...)))))))).. ( -18.22, z-score = 0.34, R) >consensus _GACGUUUUCGGGGCAUUGUAUCUGCAUUUUUAAUGCACUUCUUC__CUACCAUUAAAGGCCAAAAUGUAUUCCA_____CUUGCCCUCGAACUCGAAA_ .......(((((((((.......(((((.....)))))............((......))......................))))).))))........ (-10.52 = -11.83 + 1.31)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:54:43 2011