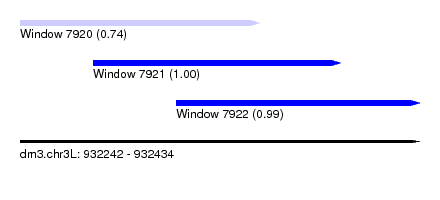
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 932,242 – 932,434 |
| Length | 192 |
| Max. P | 0.995438 |

| Location | 932,242 – 932,357 |
|---|---|
| Length | 115 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 53.06 |
| Shannon entropy | 0.67257 |
| G+C content | 0.42680 |
| Mean single sequence MFE | -31.43 |
| Consensus MFE | -10.73 |
| Energy contribution | -10.74 |
| Covariance contribution | 0.01 |
| Combinations/Pair | 1.36 |
| Mean z-score | -1.59 |
| Structure conservation index | 0.34 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.56 |
| SVM RNA-class probability | 0.744757 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
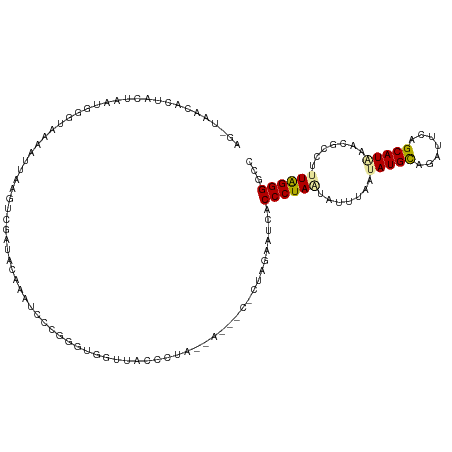
>dm3.chr3L 932242 115 + 24543557 -----AGUCGACUAAUAAGUAAACUUAGGACCACCCUAAUUCCUUAGGGUCACCCUAGUAGAUCUUUAGAUACACCCUAAUACUAAAUAUGCGAAUUCAGCAUGUACGCCUUUAGGGGUC -----.(((.((((.((((....))))((...(((((((....)))))))...)))))).)))...........((((((......((((((.......))))))......))))))... ( -29.60, z-score = -1.82, R) >droEre2.scaffold_4784 2474268 120 - 25762168 AGCUAACACUGUUAAUGGGUAAAAUUAAGUCGAUAUAAGUCCGGGGUUGUUACCCUAACAUUACACUAAUGUUACCCUAACAUUUUAUAUGCAGAUUCAGCAUAAGCGCCUCUAGGGGCC .(((.....(((((..((((((.((((.((.(((....))).(((((....)))))........)))))).))))))))))).....(((((.......))))))))((((....)))). ( -31.60, z-score = -1.51, R) >droAna3.scaffold_12948 55913 106 + 692136 AGUUAACACUUCUGAGGGGUAACAAACAGUCGGUAGAAAUCUUGGGUGAUU--------------CUUGGAUCACCCUAGUCUUUAUAAUGUCGACUCAGCAUACCCGCCUUUGGGGGCC ................(((((......(((((..(.(((.((.((((((((--------------....)))))))).))..)))....)..))))).....)))))((((....)))). ( -33.10, z-score = -1.44, R) >consensus AG_UAACACUACUAAUGGGUAAAAUUAAGUCGAUACAAAUCCCGGGUGGUUACCCUA__A___C_CUAGAAUCACCCUAAUAUUUAAUAUGCAGAUUCAGCAUAAACGCCUUUAGGGGCC ..........................................................................((((((.......(((((.......))))).......))))))... (-10.73 = -10.74 + 0.01)

| Location | 932,277 – 932,396 |
|---|---|
| Length | 119 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 52.51 |
| Shannon entropy | 0.69196 |
| G+C content | 0.47983 |
| Mean single sequence MFE | -36.03 |
| Consensus MFE | -16.91 |
| Energy contribution | -17.04 |
| Covariance contribution | 0.13 |
| Combinations/Pair | 1.41 |
| Mean z-score | -1.96 |
| Structure conservation index | 0.47 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.80 |
| SVM RNA-class probability | 0.995438 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 932277 119 + 24543557 UCCUUAGGGUCACCCUAGUAGAUCUUUAGAUACACCCUAAUACUAAAUAUGCGAAUUCAGCAUGUACGCCUUUAGGGGUCGCACUCGACUCCCAUUGGU-UAUCGAGUAUGAACUACAUA ....(((((...)))))((((.............((((((......((((((.......))))))......)))))).(((.((((((..((....)).-..)))))).))).))))... ( -34.20, z-score = -2.62, R) >droEre2.scaffold_4784 2474308 119 - 25762168 CCGGGGUUGUUACCCUAACAUUACACUAAUGUUACCCUAACAUUUUAUAUGCAGAUUCAGCAUAAGCGCCUCUAGGGGCCGUGCCUGGUUCUGAUAGGUCUCCCCAGUAUGGGGAGAUA- ..(((((....)))))(((((((...)))))))..((((.(((.....)))((((..((((((..((.((....)).)).))).)))..)))).))))(((((((.....)))))))..- ( -42.00, z-score = -2.11, R) >droAna3.scaffold_12948 55953 94 + 692136 -------------CUUGGGUGAUUCUUGGAU-CACCCUAGUCUUUAUAAUGUCGACUCAGCAUACCCGCCUUUGGGGGCCGUGCCUGGAUCCCACUGGU-UUCCG-GUAU---------- -------------...((((((((....)))-))))).((((...........))))....(((((.(((..((((..(((....)))..))))..)))-....)-))))---------- ( -31.90, z-score = -1.16, R) >consensus _C______GU_ACCCUAGCAGAUCAUUAGAU_CACCCUAAUAUUUAAUAUGCAGAUUCAGCAUAAACGCCUUUAGGGGCCGUGCCUGGAUCCCAUUGGU_UACCGAGUAUG_________ ..................................((((((.......(((((.......))))).......)))))).((((((.(((..((....))....))).))))))........ (-16.91 = -17.04 + 0.13)

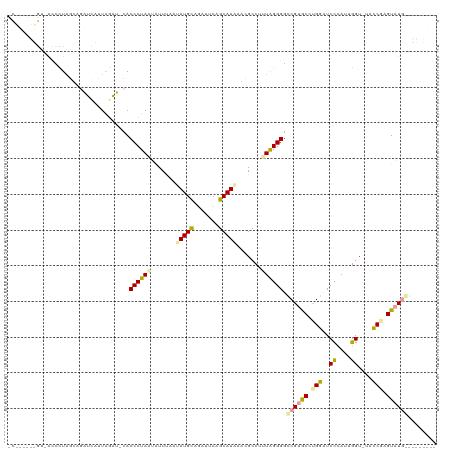
| Location | 932,317 – 932,434 |
|---|---|
| Length | 117 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 52.59 |
| Shannon entropy | 0.65035 |
| G+C content | 0.49331 |
| Mean single sequence MFE | -35.47 |
| Consensus MFE | -13.56 |
| Energy contribution | -12.90 |
| Covariance contribution | -0.66 |
| Combinations/Pair | 1.40 |
| Mean z-score | -2.28 |
| Structure conservation index | 0.38 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.20 |
| SVM RNA-class probability | 0.985375 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
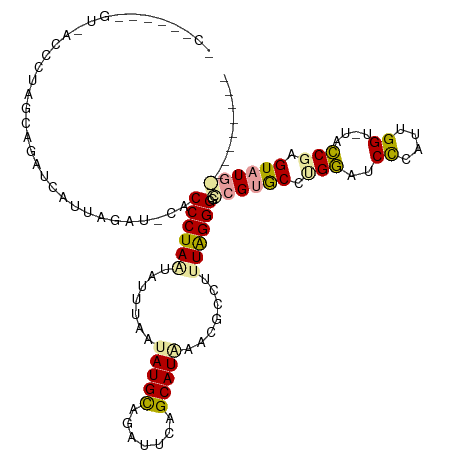
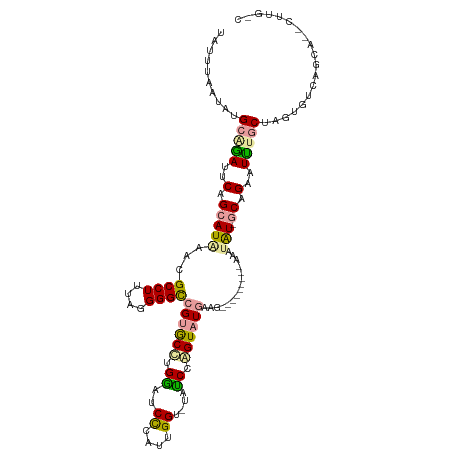
>dm3.chr3L 932317 117 + 24543557 UACUAAAUAUGCGAAUUCAGCAUGUACGCCUUUAGGGGUCGCACUCGACUCCCAUUGGU-UAUCGAGUAUGAACUACAUACUUACAUAUUGCAGAAUUUGCUAGUGUCAGCA--CUUGGC ..((((....((((((((.((((((..(((..(.(((((((....))))))).)..)))-....(((((((.....))))))))))...))).))))))))..(((....))--))))). ( -35.30, z-score = -2.87, R) >droEre2.scaffold_4784 2474348 110 - 25762168 CAUUUUAUAUGCAGAUUCAGCAUAAGCGCCUCUAGGGGCCGUGCCUGGUUCUGAUAGGUCUCCCCAGUAUGGGG---------AGAUAU-GCUGUUUCUGCUUGUGUCAGCAGUCUUGCC .....((((.(((((..(((((((...((((...(((((((....)))))))...))))((((((.....))))---------)).)))-))))..))))).))))...((......)). ( -44.80, z-score = -3.19, R) >droAna3.scaffold_12948 55979 92 + 692136 UCUUUAUAAUGUCGACUCAGCAUACCCGCCUUUGGGGGCCGUGCCUGGAUCCCACUGGU--UUCCGGUAUCUUGC--------GAUCGU--CAGCAUUUCCCAC---------------- .......(((((.(((...(((((((.(((..((((..(((....)))..))))..)))--....))))...)))--------....))--).)))))......---------------- ( -26.30, z-score = -0.79, R) >consensus UAUUUAAUAUGCAGAUUCAGCAUAAACGCCUUUAGGGGCCGUGCCUGGAUCCCAUUGGU_UAUCCAGUAUGAAG_________AAAUAU_GCAGAAUUUGCUAGUGUCAGCA__CUUG_C .......(((((.......)))))...(((....(((((((....)))))))....)))............................................................. (-13.56 = -12.90 + -0.66)

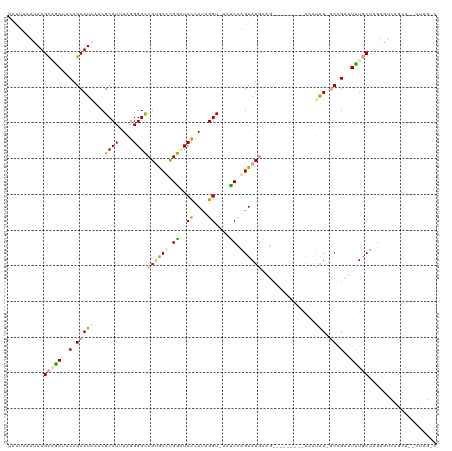
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:54:39 2011