| Sequence ID | dm3.chr2L |
|---|---|
| Location | 4,086,927 – 4,087,056 |
| Length | 129 |
| Max. P | 0.902246 |

| Location | 4,086,927 – 4,087,056 |
|---|---|
| Length | 129 |
| Sequences | 7 |
| Columns | 144 |
| Reading direction | forward |
| Mean pairwise identity | 81.95 |
| Shannon entropy | 0.34590 |
| G+C content | 0.39878 |
| Mean single sequence MFE | -32.12 |
| Consensus MFE | -16.93 |
| Energy contribution | -17.93 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.22 |
| Structure conservation index | 0.53 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.62 |
| SVM RNA-class probability | 0.763633 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
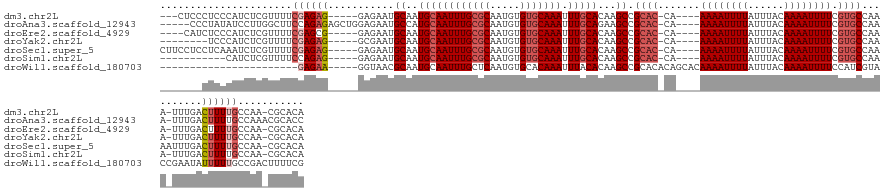
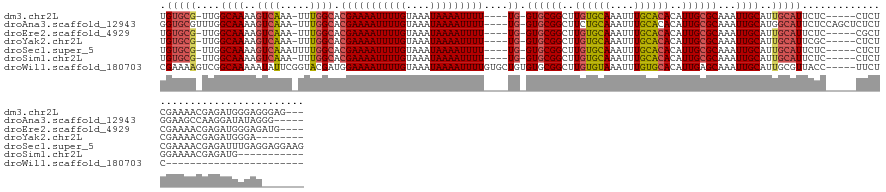
>dm3.chr2L 4086927 129 + 23011544 ---CUCCCUCCCAUCUCGUUUUCGAGAG-----GAGAAUGCAAUGCAAUUUGCGCAAUGUGUGCAAAUUUGCACAAGCCGCAC-CA----AAAAUUUUAUUUACAAAAUUUUCGUGCCAAA-UUUGACUUUUGCCAA-CGCACA ---....((((..(((((....))))))-----)))..(((..((((((((((((.....))))))).)))))((((..((((-..----((((((((......)))))))).))))....-))))...........-.))).. ( -35.40, z-score = -3.16, R) >droAna3.scaffold_12943 653560 133 - 5039921 -----CCCUAUAUCCUUGGCUUCCAGAGAGCUGGAGAAUGCCAUGCAAUUUGCGCAAUGUGUGCAAAUUUGCAGAAGCCGCAC-CA----AAAAUUUUAUUUACAAAAUUUUCGUGCCAAA-UUUGACUUUUGCCAAACGCACC -----...........((((((((((....))))))...))))((((((((((((.....))))))))).(((((((..((((-..----((((((((......)))))))).))))((..-..)).))))))).....))).. ( -37.00, z-score = -2.76, R) >droEre2.scaffold_4929 4144918 128 + 26641161 ----CAUCUCCCAUCUCGUUUUCGAGCG-----GAGAAUGCAAUGCAAUUUGCGCAAUGUGUGCAAAUUUGCACAAGCCGCAC-CA----AAAAUUUUAUUUACAAAAUUUUCGUGCCAAA-UUUGACUUUUGCCAA-CGCACA ----..(((((...((((....)))).)-----)))).(((..((((((((((((.....))))))).)))))((((..((((-..----((((((((......)))))))).))))....-))))...........-.))).. ( -35.20, z-score = -2.64, R) >droYak2.chr2L 4104051 124 + 22324452 --------UCCCAUCUCGUUUUCGAGAG-----GCGAAUGCAAUGCAAUUUGCGCAAUGUGUGCAAAUUUGCACAAGCCGCAC-CA----AAAAUUUUAUUUACAAAAUUUUCGUGCCAAA-UUUGACUUUUGCCAA-CGCACA --------.....(((((....)))))(-----(((((..(((((((((((((((.....))))))).)))))......((((-..----((((((((......)))))))).))))....-.)))...))))))..-...... ( -36.20, z-score = -2.94, R) >droSec1.super_5 2186073 133 + 5866729 CUUCCUCCUCAAAUCUCGUUUUCGAGAG-----GAGAAUGCAAUGCAAUUUGCGCAAUGUGUGCAAAUUUGCACAAGCCGCAC-CA----AAAAUUUUAUUUACAAAAUUUUCGUGCCAAAAUUUGACUUUUGCCAA-CGCACA .......(((...(((((....))))).-----)))..(((..((((((((((((.....))))))).)))))......((((-..----((((((((......)))))))).))))....................-.))).. ( -33.80, z-score = -2.35, R) >droSim1.chr2L 4042087 121 + 22036055 -----------CAUCUCGUUUUCCAGAG-----GAGAAUGCAAUGCAAUUUGCGCAAUGUGUGCAAAUUUGCACAAGCCGCAC-CA----AAAAUUUUAUUUACAAAAUUUUCGUGCCAAA-UUUGACUUUUGCCAA-CGCACA -----------..((((.((.....)).-----)))).(((..((((((((((((.....))))))).)))))((((..((((-..----((((((((......)))))))).))))....-))))...........-.))).. ( -29.10, z-score = -1.17, R) >droWil1.scaffold_180703 2081315 116 + 3946847 -----------------------GAGAA-----GGUAACGCAAUGCAAUUUGCUCAAUGUGCACAAAUUUACACAAGCCGCACACAGCACAAAAUUUUAUUUACAAAAUUUUCCAUCGUACCGAAUAUUUUUGCCGACUUUUCG -----------------------(((((-----(.....((((.......((((...(((((.................))))).)))).((((((((......))))))))..................))))...)))))). ( -18.13, z-score = -0.54, R) >consensus _______CUCCCAUCUCGUUUUCGAGAG_____GAGAAUGCAAUGCAAUUUGCGCAAUGUGUGCAAAUUUGCACAAGCCGCAC_CA____AAAAUUUUAUUUACAAAAUUUUCGUGCCAAA_UUUGACUUUUGCCAA_CGCACA .......................................((((((((((((((((.....))))))).)))))......((((.......((((((((......)))))))).)))).............)))).......... (-16.93 = -17.93 + 1.00)
| Location | 4,086,927 – 4,087,056 |
|---|---|
| Length | 129 |
| Sequences | 7 |
| Columns | 144 |
| Reading direction | reverse |
| Mean pairwise identity | 81.95 |
| Shannon entropy | 0.34590 |
| G+C content | 0.39878 |
| Mean single sequence MFE | -39.06 |
| Consensus MFE | -22.92 |
| Energy contribution | -23.36 |
| Covariance contribution | 0.43 |
| Combinations/Pair | 1.15 |
| Mean z-score | -2.27 |
| Structure conservation index | 0.59 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.16 |
| SVM RNA-class probability | 0.902246 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
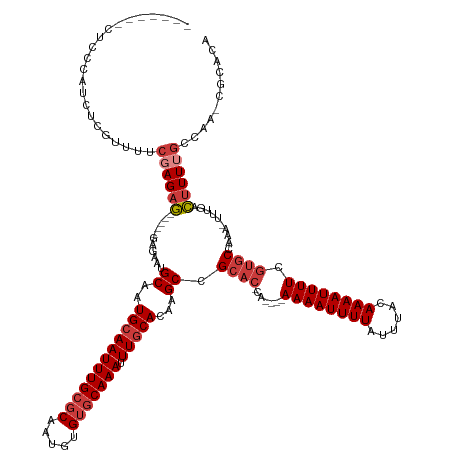
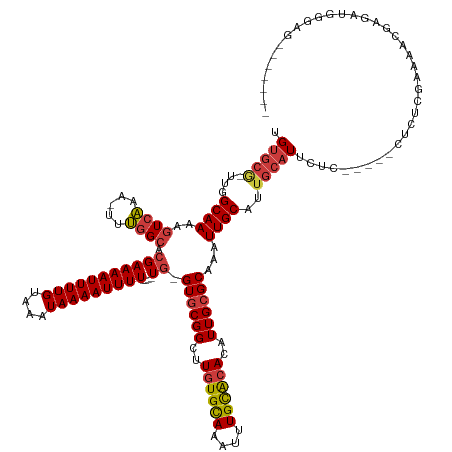
>dm3.chr2L 4086927 129 - 23011544 UGUGCG-UUGGCAAAAGUCAAA-UUUGGCACGAAAAUUUUGUAAAUAAAAUUUU----UG-GUGCGGCUUGUGCAAAUUUGCACACAUUGCGCAAAUUGCAUUGCAUUCUC-----CUCUCGAAAACGAGAUGGGAGGGAG--- .(((((-...((((..((((..-..)))).(((((((((((....)))))))))----))-((((((..((((((....))))))..))))))...))))..)))))((((-----((((((....)))))..)))))...--- ( -44.90, z-score = -3.73, R) >droAna3.scaffold_12943 653560 133 + 5039921 GGUGCGUUUGGCAAAAGUCAAA-UUUGGCACGAAAAUUUUGUAAAUAAAAUUUU----UG-GUGCGGCUUCUGCAAAUUUGCACACAUUGCGCAAAUUGCAUGGCAUUCUCCAGCUCUCUGGAAGCCAAGGAUAUAGGG----- .(((((((((((....))))))-)...))))..................(((((----((-((...(((..(((((.(((((.(.....).)))))))))).)))....(((((....))))).)))))))))......----- ( -38.50, z-score = -1.29, R) >droEre2.scaffold_4929 4144918 128 - 26641161 UGUGCG-UUGGCAAAAGUCAAA-UUUGGCACGAAAAUUUUGUAAAUAAAAUUUU----UG-GUGCGGCUUGUGCAAAUUUGCACACAUUGCGCAAAUUGCAUUGCAUUCUC-----CGCUCGAAAACGAGAUGGGAGAUG---- .(((((-...((((..((((..-..)))).(((((((((((....)))))))))----))-((((((..((((((....))))))..))))))...))))..)))))((((-----(.((((....))))...)))))..---- ( -44.90, z-score = -3.50, R) >droYak2.chr2L 4104051 124 - 22324452 UGUGCG-UUGGCAAAAGUCAAA-UUUGGCACGAAAAUUUUGUAAAUAAAAUUUU----UG-GUGCGGCUUGUGCAAAUUUGCACACAUUGCGCAAAUUGCAUUGCAUUCGC-----CUCUCGAAAACGAGAUGGGA-------- .(((((-...((((..((((..-..)))).(((((((((((....)))))))))----))-((((((..((((((....))))))..))))))...))))..)))))((.(-----((((((....))))).))))-------- ( -41.60, z-score = -2.68, R) >droSec1.super_5 2186073 133 - 5866729 UGUGCG-UUGGCAAAAGUCAAAUUUUGGCACGAAAAUUUUGUAAAUAAAAUUUU----UG-GUGCGGCUUGUGCAAAUUUGCACACAUUGCGCAAAUUGCAUUGCAUUCUC-----CUCUCGAAAACGAGAUUUGAGGAGGAAG ..((((-...((((..((((.....)))).(((((((((((....)))))))))----))-((((((..((((((....))))))..))))))...))))..))))(((((-----((((((....))).....)))))))).. ( -44.20, z-score = -3.36, R) >droSim1.chr2L 4042087 121 - 22036055 UGUGCG-UUGGCAAAAGUCAAA-UUUGGCACGAAAAUUUUGUAAAUAAAAUUUU----UG-GUGCGGCUUGUGCAAAUUUGCACACAUUGCGCAAAUUGCAUUGCAUUCUC-----CUCUGGAAAACGAGAUG----------- .(((((-...((((..((((..-..)))).(((((((((((....)))))))))----))-((((((..((((((....))))))..))))))...))))..)))))....-----(((........)))...----------- ( -35.30, z-score = -1.66, R) >droWil1.scaffold_180703 2081315 116 - 3946847 CGAAAAGUCGGCAAAAAUAUUCGGUACGAUGGAAAAUUUUGUAAAUAAAAUUUUGUGCUGUGUGCGGCUUGUGUAAAUUUGUGCACAUUGAGCAAAUUGCAUUGCGUUACC-----UUCUC----------------------- (((....)))............((((((....(((((((((....)))))))))((((....(((....((((((......))))))....)))....))))..)).))))-----.....----------------------- ( -24.00, z-score = 0.32, R) >consensus UGUGCG_UUGGCAAAAGUCAAA_UUUGGCACGAAAAUUUUGUAAAUAAAAUUUU____UG_GUGCGGCUUGUGCAAAUUUGCACACAUUGCGCAAAUUGCAUUGCAUUCUC_____CUCUCGAAAACGAGAUGGGAG_______ .(((((....((((..((((.....))))...(((((((((....))))))))).......((((((..((((((....))))))..))))))...))))..)))))..................................... (-22.92 = -23.36 + 0.43)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:15:20 2011