| Sequence ID | dm3.chr3L |
|---|---|
| Location | 923,029 – 923,125 |
| Length | 96 |
| Max. P | 0.952008 |

| Location | 923,029 – 923,125 |
|---|---|
| Length | 96 |
| Sequences | 9 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 77.10 |
| Shannon entropy | 0.43149 |
| G+C content | 0.48864 |
| Mean single sequence MFE | -24.84 |
| Consensus MFE | -15.81 |
| Energy contribution | -16.49 |
| Covariance contribution | 0.68 |
| Combinations/Pair | 1.16 |
| Mean z-score | -1.89 |
| Structure conservation index | 0.64 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.58 |
| SVM RNA-class probability | 0.952008 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
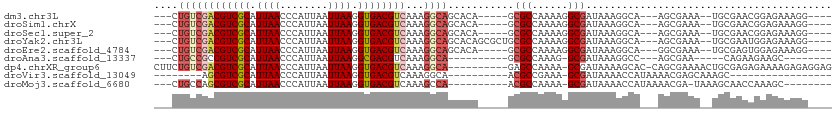
>dm3.chr3L 923029 96 - 24543557 ---CUGUCGACGUCGCAUUAACCCAUUAAUUAAGGUGACGUCAAAGGCAGCACA-----GCGCCAAAAGGCGAUAAAGGCA---AGCGAAA--UGCGAACGGAGAAAGG---- ---(((((((((((((.((((........)))).))))))))....((.((...-----.((((....))))......)).---.))....--...).)))).......---- ( -26.60, z-score = -1.74, R) >droSim1.chrX 9375325 96 - 17042790 ---CUGUCGACGUCGCAUUAACCCAUUAAUUAAGGUGACGUCAAAGGCAGCACA-----GCGCCAAAAGGCGAUAAAGGCA---AGCGAAA--UGCGAACGGAGAAAGG---- ---(((((((((((((.((((........)))).))))))))....((.((...-----.((((....))))......)).---.))....--...).)))).......---- ( -26.60, z-score = -1.74, R) >droSec1.super_2 942143 96 - 7591821 ---CUGUCGACGUCGCAUUAACCCAUUAAUUAAGGUGACGUCAAAGGCAGCACA-----GCGCCAAAAGGCGAUAAAGGCA---AGCGAAA--UGCGAACGGAGAAAGG---- ---(((((((((((((.((((........)))).))))))))....((.((...-----.((((....))))......)).---.))....--...).)))).......---- ( -26.60, z-score = -1.74, R) >droYak2.chr3L 904155 101 - 24197627 ---CUGUCGACGUCGCAUUAACCCAUUAAUUAAGGUGACGUCAAAGGCAGCACAGCGCUGCGCCAAAAGGCGAUAAAGGCA---AGCGAAA--UGCGAAUGGAGAAAGG---- ---.((((((((((((.((((........)))).))))))))....(((((.....)))))(((....)))......))))---.((....--.)).............---- ( -31.40, z-score = -2.03, R) >droEre2.scaffold_4784 933042 96 - 25762168 ---CUGUCGACGUCGCAUUAACCCAUUAAUUAAGGUGACGUCAAAGGCAGCACA-----GCGCCAAAAGGCGAUAAAGGCA---GGCGAAA--UGCGAGUGGAGAAAGG---- ---(((((((((((((.((((........)))).))))))))...)))))(((.-----.((((....))))......(((---.......--)))..)))........---- ( -29.20, z-score = -2.24, R) >droAna3.scaffold_13337 16304268 83 + 23293914 ---CUGCCGCCGUCGCAUUAACCCAUUAAUUAAGGCGACGUCAAAGGCA----------GCGCCAAAG-GCGAUAAAGGCC---AGCGAA-----CAGAAGAAGC-------- ---(((.(((((((((.((((........)))).)))))).....(((.----------.((((...)-)))......)))---.)))..-----))).......-------- ( -25.30, z-score = -1.91, R) >dp4.chrXR_group6 6544119 101 - 13314419 CUUCUGUCGACGUCGCAUUAACCCAUUAAUUAAGGUGACGUCAAAGGCA----------GAGCCAAAA-GCGAUAAAAGCAC-CAGCGAAAACUGCGAGAGAAAAGAGAGGAG (((((.((((((((((.((((........)))).))))))))....(((----------(.((.....-)).......((..-..)).....)))).........)).))))) ( -29.30, z-score = -3.50, R) >droVir3.scaffold_13049 15480362 77 - 25233164 --------AGCGUCGCAUUAACCCAUUAAUUAAGGUGACGUCAAAGGCA----------ACGCCGAAA-GCGAUAAAACCAUAAAACGAGCAAAGC----------------- --------.(((((((.((((........)))).)))))(((...(...----------.)(.(....-)))))...............)).....----------------- ( -13.70, z-score = -0.66, R) >droMoj3.scaffold_6680 3391432 90 + 24764193 ---CUGCCAGCGUCGCAUUAACCCAUUAAUUAAGGUGACGUCAAAGCCA----------ACGCCAAAA-GCGAUAAAACCAUAAAACGA-UAAAGCAACCAAAGC-------- ---.(((..(((((((.((((........)))).)))))))........----------.(((.....-))).................-....)))........-------- ( -14.90, z-score = -1.44, R) >consensus ___CUGUCGACGUCGCAUUAACCCAUUAAUUAAGGUGACGUCAAAGGCAGCACA_____GCGCCAAAAGGCGAUAAAGGCA___AGCGAAA__UGCGAACGGAGAAAGG____ ....((((((((((((.((((........)))).))))))))...))))...........(((......)))......................................... (-15.81 = -16.49 + 0.68)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:54:37 2011