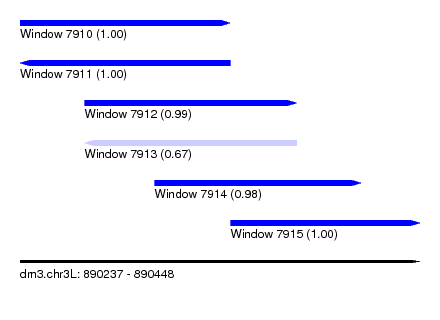
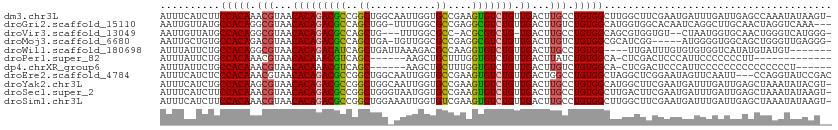
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 890,237 – 890,448 |
| Length | 211 |
| Max. P | 0.999551 |

| Location | 890,237 – 890,348 |
|---|---|
| Length | 111 |
| Sequences | 11 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 65.38 |
| Shannon entropy | 0.77724 |
| G+C content | 0.49801 |
| Mean single sequence MFE | -31.29 |
| Consensus MFE | -21.03 |
| Energy contribution | -20.10 |
| Covariance contribution | -0.93 |
| Combinations/Pair | 1.41 |
| Mean z-score | -1.81 |
| Structure conservation index | 0.67 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 4.00 |
| SVM RNA-class probability | 0.999551 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
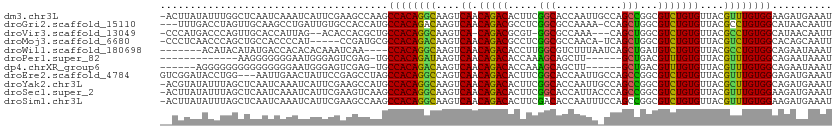
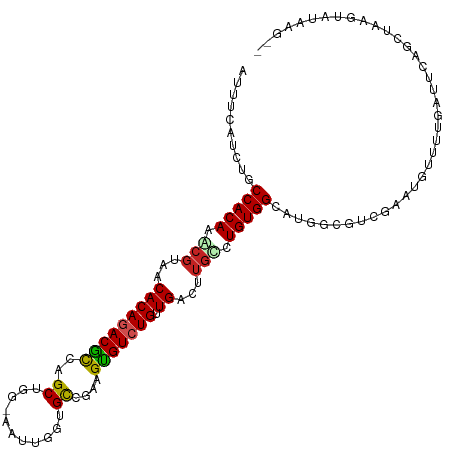
>dm3.chr3L 890237 111 + 24543557 -ACUUAUAUUUGGCUCAAUCAAAUCAUUCGAAGCCAAGCCACAGGCAAGUCAACAGACACUUCGGCACCAAUUGCCAGCCGGCGUCUGUGUUACGUUUGUGGAAGAUGAAAU -.(((...(((((((((((......))).).)))))))((((((((......((((((.((..((((.....))))....)).)))))).....)))))))))))....... ( -32.00, z-score = -1.96, R) >droGri2.scaffold_15110 17215499 108 + 24565398 ---UUUGACCUAGUUGCAAGCCUGAUUGUGCCACCAUGCCACAGACAAGUCAACAGACGCCUCGGCGCCAAAA-CCAGCUGGCGUCUGUGUUACGCCUGUGGCAUAACAAUU ---.(((.....(..((((......))))..)...(((((((((....((..(((((((((..(((.......-...))))))))))))...))..)))))))))..))).. ( -37.60, z-score = -2.44, R) >droVir3.scaffold_13049 15442463 104 + 25233164 -CCCAUGACCCAGUUGCACCAUUAG--ACACCACGCUGCCACAGGCAAGUCA-CAGACGCGU-GGCGCCAAA---CAGCUGGCGUCUGUGUUACGCCUGUGGCAUAACAAUU -........................--.........((((((((((....((-(((((((.(-(((......---..)))))))))))))....))))))))))........ ( -41.60, z-score = -3.15, R) >droMoj3.scaffold_6680 3348583 105 - 24764193 -CCCUCAACCCAGCUGCCACCCCAU-----CCGAUGCGCCACAGACAAGUCAACAGACGCCUCGGCGCCAACA-UCAGCUGGCGUCUGUGUUACGUCUGUGGCACAGCAAUU -...........((((......(((-----...))).(((((((((......(((((((((..(((.......-...)))))))))))).....))))))))).)))).... ( -40.60, z-score = -4.14, R) >droWil1.scaffold_180698 5597256 101 + 11422946 -------ACAUACAUAUGACCACACACAAAUCAA----CCACAGGCAAGUCAACAGACACCUUGGCGUCUUUAAUCAGCUGAUGUCUGUGUUACGCCUGUGGCAGAAUAAAU -------.......................((..----((((((((......(((((((..(..((...........))..)))))))).....))))))))..))...... ( -29.50, z-score = -2.97, R) >droPer1.super_82 161566 92 - 267963 -------------AAGGGGGGGAAUGGGAGUCGAG-UGCCACAGAUAAGUCAACAGACACCAAAGCAGCUU------GCUGACGUUUGUGUUACGUUUGUGGCAGAAUAAAU -------------......................-((((((((((......((((((.....(((.....------)))...)))))).....))))))))))........ ( -22.80, z-score = -0.38, R) >dp4.chrXR_group6 6491430 99 + 13314419 ------AGGGGGGGGGGGGGGGAAUGGGAGUCGAG-UGCCACAGACAAGUCAACAGACACCAAAGCAGCUU------GCUGACGUUUGUGUUACGUUUGUGGCAGAAUAAAU ------.............................-((((((((((......((((((.....(((.....------)))...)))))).....))))))))))........ ( -24.30, z-score = 0.12, R) >droEre2.scaffold_4784 897304 109 + 25762168 GUCGGAUACCUGG---AAUUGAACUAUUCCGAGCCUAGCCACAGGCCAGUCAACAGACACUUCGGCACCAAUUGCCAGCCGGCGUCUGUGUUACGUUUGUGGGAGAUGAAAU (((((.....(((---(((......))))))..))...((((((((......((((((.((..((((.....))))....)).)))))).....))))))))..)))..... ( -32.60, z-score = -0.66, R) >droYak2.chr3L 869510 111 + 24197627 -ACGUAUAUUUAGCUCAAUCAAAUCAUUCGAAGCCAUGCCACAGGCAAGUCAACAGACACUUCGGCACCAAUUGCCAGCCGGCGUCUGUGUUACGCUUGUGGCAGAUGAAAU -......................(((((........((((((((((......((((((.((..((((.....))))....)).)))))).....)))))))))))))))... ( -33.60, z-score = -2.20, R) >droSec1.super_2 909806 111 + 7591821 -ACUUAUAUUUAGCUCAAUCAAAUCAUUCGAAGUCAAGCCACAGGCAAGUCAACAGACACUUCGGCACCAUUACCCAGCCGGCGUCUGUGUUACGUUUGUGGAAGAUGAAAU -...............................(((...((((((((......((((((...(((((...........))))).)))))).....))))))))..)))..... ( -26.80, z-score = -1.55, R) >droSim1.chr3L 532913 111 + 22553184 -ACUUAUAUUUAGCUCAAUCAAAUCAUUCGAAGCCAAGCCACAGGCAAGUCAACAGACACUUCGACACCAAUUUCCAGCCGGCGUCUGUGUUACGUUUGUGGAAGAUGAAAU -......................((((((((((....(((...)))..(((....))).))))))......((((((.(..((((.......))))..)))))))))))... ( -22.80, z-score = -0.53, R) >consensus __CUUAUACCUAGCUGAAUCAAAACAUUCGACGCCAUGCCACAGGCAAGUCAACAGACACCUCGGCACCAAUA_CCAGCUGGCGUCUGUGUUACGUUUGUGGCAGAAGAAAU ......................................((((((((......((((((.....(((...........)))...)))))).....)))))))).......... (-21.03 = -20.10 + -0.93)
| Location | 890,237 – 890,348 |
|---|---|
| Length | 111 |
| Sequences | 11 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 65.38 |
| Shannon entropy | 0.77724 |
| G+C content | 0.49801 |
| Mean single sequence MFE | -34.04 |
| Consensus MFE | -16.48 |
| Energy contribution | -16.45 |
| Covariance contribution | -0.03 |
| Combinations/Pair | 1.32 |
| Mean z-score | -1.81 |
| Structure conservation index | 0.48 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.17 |
| SVM RNA-class probability | 0.997773 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 890237 111 - 24543557 AUUUCAUCUUCCACAAACGUAACACAGACGCCGGCUGGCAAUUGGUGCCGAAGUGUCUGUUGACUUGCCUGUGGCUUGGCUUCGAAUGAUUUGAUUGAGCCAAAUAUAAGU- ..........(((((...((((.((((((((((((...........))))..))))))))....)))).))))).((((((((((.....))))...))))))........- ( -30.90, z-score = -0.87, R) >droGri2.scaffold_15110 17215499 108 - 24565398 AAUUGUUAUGCCACAGGCGUAACACAGACGCCAGCUGG-UUUUGGCGCCGAGGCGUCUGUUGACUUGUCUGUGGCAUGGUGGCACAAUCAGGCUUGCAACUAGGUCAAA--- ..(..((((((((((((((....(((((((((...(((-(......)))).))))))))).....))))))))))))))..)........((((((....))))))...--- ( -47.30, z-score = -3.30, R) >droVir3.scaffold_13049 15442463 104 - 25233164 AAUUGUUAUGCCACAGGCGUAACACAGACGCCAGCUG---UUUGGCGCC-ACGCGUCUG-UGACUUGCCUGUGGCAGCGUGGUGU--CUAAUGGUGCAACUGGGUCAUGGG- ...(((((((((...)))))))))(((((.((((.((---(..((((((-(((((((..-.))).((((...)))))))))))))--).......))).))))))).))..- ( -46.20, z-score = -2.26, R) >droMoj3.scaffold_6680 3348583 105 + 24764193 AAUUGCUGUGCCACAGACGUAACACAGACGCCAGCUGA-UGUUGGCGCCGAGGCGUCUGUUGACUUGUCUGUGGCGCAUCGG-----AUGGGGUGGCAGCUGGGUUGAGGG- ....(((((((((((((((....((((((((((((...-.))))))((....)))))))).....)))))))))).((((..-----....)))))))))...........- ( -50.70, z-score = -3.53, R) >droWil1.scaffold_180698 5597256 101 - 11422946 AUUUAUUCUGCCACAGGCGUAACACAGACAUCAGCUGAUUAAAGACGCCAAGGUGUCUGUUGACUUGCCUGUGG----UUGAUUUGUGUGUGGUCAUAUGUAUGU------- ......((.((((((((((....(((((((((.((...........))...))))))))).....)))))))))----).))...(((((....)))))......------- ( -34.10, z-score = -2.30, R) >droPer1.super_82 161566 92 + 267963 AUUUAUUCUGCCACAAACGUAACACAAACGUCAGC------AAGCUGCUUUGGUGUCUGUUGACUUAUCUGUGGCA-CUCGACUCCCAUUCCCCCCCUU------------- ........(((((((...((.......))((((((------(..(.((....)))..))))))).....)))))))-......................------------- ( -20.80, z-score = -2.29, R) >dp4.chrXR_group6 6491430 99 - 13314419 AUUUAUUCUGCCACAAACGUAACACAAACGUCAGC------AAGCUGCUUUGGUGUCUGUUGACUUGUCUGUGGCA-CUCGACUCCCAUUCCCCCCCCCCCCCCCU------ ........(((((((.((((.......))((((((------(..(.((....)))..)))))))..)).)))))))-.............................------ ( -22.60, z-score = -2.89, R) >droEre2.scaffold_4784 897304 109 - 25762168 AUUUCAUCUCCCACAAACGUAACACAGACGCCGGCUGGCAAUUGGUGCCGAAGUGUCUGUUGACUGGCCUGUGGCUAGGCUCGGAAUAGUUCAAUU---CCAGGUAUCCGAC .......................((((((((((((...........))))..))))))))...((((((...))))))((..(((((......)))---))..))....... ( -31.50, z-score = -0.15, R) >droYak2.chr3L 869510 111 - 24197627 AUUUCAUCUGCCACAAGCGUAACACAGACGCCGGCUGGCAAUUGGUGCCGAAGUGUCUGUUGACUUGCCUGUGGCAUGGCUUCGAAUGAUUUGAUUGAGCUAAAUAUACGU- ........(((((((.(((....((((((((((((...........))))..)))))))).....))).)))))))(((((((((.....))))...))))).........- ( -36.90, z-score = -1.95, R) >droSec1.super_2 909806 111 - 7591821 AUUUCAUCUUCCACAAACGUAACACAGACGCCGGCUGGGUAAUGGUGCCGAAGUGUCUGUUGACUUGCCUGUGGCUUGACUUCGAAUGAUUUGAUUGAGCUAAAUAUAAGU- .......................((((((((((((...........))))..))))))))..(((((....(((((..(..((((.....)))))..)))))....)))))- ( -27.50, z-score = -0.45, R) >droSim1.chr3L 532913 111 - 22553184 AUUUCAUCUUCCACAAACGUAACACAGACGCCGGCUGGAAAUUGGUGUCGAAGUGUCUGUUGACUUGCCUGUGGCUUGGCUUCGAAUGAUUUGAUUGAGCUAAAUAUAAGU- ..........(((((...((((.((((((((((((...........))))..))))))))....)))).))))).((((((((((.....))))...))))))........- ( -25.90, z-score = 0.04, R) >consensus AUUUCAUCUGCCACAAACGUAACACAGACGCCAGCUGG_AAUUGGUGCCGAAGUGUCUGUUGACUUGCCUGUGGCAUGGCGUCGAAUGUUUUGAUUCAGCUAAGUAUAAG__ ..........(((((.(((...(((((((((..((...........))....))))))).))...))).)))))...................................... (-16.48 = -16.45 + -0.03)
| Location | 890,271 – 890,383 |
|---|---|
| Length | 112 |
| Sequences | 12 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 70.64 |
| Shannon entropy | 0.63498 |
| G+C content | 0.54318 |
| Mean single sequence MFE | -37.03 |
| Consensus MFE | -22.84 |
| Energy contribution | -22.25 |
| Covariance contribution | -0.59 |
| Combinations/Pair | 1.47 |
| Mean z-score | -1.37 |
| Structure conservation index | 0.62 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.33 |
| SVM RNA-class probability | 0.988590 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
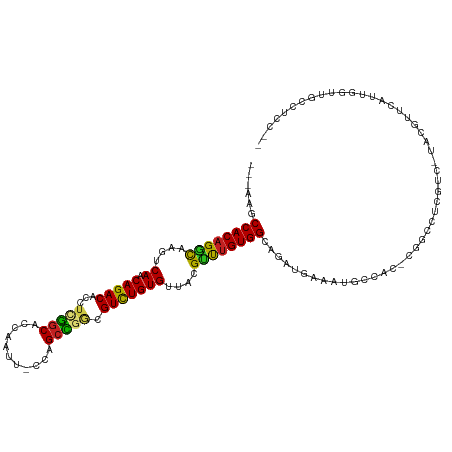
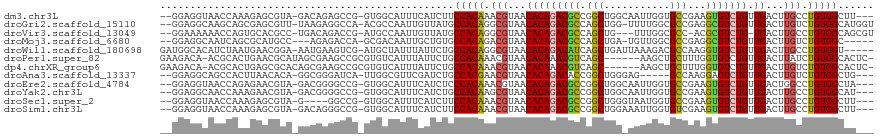
>dm3.chr3L 890271 112 + 24543557 ---AAGCCACAGGCAAGUCAACAGACACUUCGGCACCAAUUGCCAGCCGGCGUCUGUGUUACGUUUGUGGAAGAUGAAAUGCCAC-CGGCUCUGUC-UACGCUCUUUGGUUACCUCC-- ---.(((((.((((..((..(((((......((((.....)))).(((((.(...(((((.(((((.....))))).)))))).)-))))))))).-.)))).)).)))))......-- ( -34.30, z-score = -0.75, R) >droGri2.scaffold_15110 17215528 114 + 24565398 ACCAUGCCACAGACAAGUCAACAGACGCCUCGGCGCCAAAA-CCAGCUGGCGUCUGUGUUACGCCUGUGGCAUAACAAUUGGCGU-UGGCCUCUUA-AACGCUCGCUGCUUGCCUCC-- ...(((((((((....((..(((((((((..(((.......-...))))))))))))...))..))))))))).......(((((-(.........-)))))).((.....))....-- ( -41.30, z-score = -2.38, R) >droVir3.scaffold_13049 15442492 110 + 25233164 ACGCUGCCACAGGCAAGUCA-CAGACGCGU-GGCGCCAAA---CAGCUGGCGUCUGUGUUACGCCUGUGGCAUAACAAUUGGCAU-CGGUCUGUCA-GGCGUGCACUGGUUUUUUCC-- ((((((((((((((....((-(((((((.(-(((......---..)))))))))))))....)))))))))........(((((.-.....)))))-)))))...............-- ( -50.80, z-score = -3.52, R) >droMoj3.scaffold_6680 3348614 107 - 24764193 -----GCCACAGACAAGUCAACAGACGCCUCGGCGCCAACA-UCAGCUGGCGUCUGUGUUACGUCUGUGGCACAGCAAUUGUCGC-UGGUCUCU---GGCAUGCGCUGAUUGCCUCC-- -----(((((((((......(((((((((..(((.......-...)))))))))))).....)))))))))...(((((((.(((-..((....---.))..))).)))))))....-- ( -48.30, z-score = -3.58, R) >droWil1.scaffold_180698 5597282 112 + 11422946 -----ACCACAGGCAAGUCAACAGACACCUUGGCGUCUUUAAUCAGCUGAUGUCUGUGUUACGCCUGUGGCAGAAUAAAUAGCAU-CGACUUCAUU-UCCGUUCAUUAGAUGUGCCAUC -----.((((((((......(((((((..(..((...........))..)))))))).....))))))))...........((((-(.........-...........)))))...... ( -32.05, z-score = -2.04, R) >droPer1.super_82 161585 111 - 267963 -GAGUGCCACAGAUAAGUCAACAGACACCAAAGCAGCUU------GCUGACGUUUGUGUUACGUUUGUGGCAGAAUAAAUGACACGCGGCUUCGCUAUGCGUUCAGUGCGU-UGUCUUC -...((((((((((......((((((.....(((.....------)))...)))))).....))))))))))........((((((((.((.(((...)))...)))))).-))))... ( -34.50, z-score = -0.57, R) >dp4.chrXR_group6 6491456 111 + 13314419 -GAGUGCCACAGACAAGUCAACAGACACCAAAGCAGCUU------GCUGACGUUUGUGUUACGUUUGUGGCAGAAUAAAUGACACGCGGCUUCGCUGUGCGCUCAGUGCGU-UGUCUUC -...((((((((((......((((((.....(((.....------)))...)))))).....))))))))))........((((((((((...))))))(((.....))).-))))... ( -36.80, z-score = -0.30, R) >droAna3.scaffold_13337 16266106 107 - 23293914 ---CAGCCACAGACAAGUCAACAGACUCCUUGGC-----CUCCCAGCCGGUGUCUGUGUUACGUUCGUGGCAGAUCGAACGCCAA-UGAUCCCGCC-UGUGUUAAGUGGCUGCCUCC-- ---(((((((.((((.((..((((((.((..(((-----......))))).))))))...))......(((.(((((........-)))))..)))-..))))..))))))).....-- ( -37.20, z-score = -2.16, R) >droEre2.scaffold_4784 897336 112 + 25762168 ---UAGCCACAGGCCAGUCAACAGACACUUCGGCACCAAUUGCCAGCCGGCGUCUGUGUUACGUUUGUGGGAGAUGAAAUGCCAC-CGGCCCCGUC-UACGUUCUCUGGUUACCUCC-- ---........((((((..(((((((.....((((.....)))).(((((.(...(((((.(((((.....))))).)))))).)-))))...)))-)..)))..))))))......-- ( -33.00, z-score = 0.08, R) >droYak2.chr3L 869544 112 + 24197627 ---AUGCCACAGGCAAGUCAACAGACACUUCGGCACCAAUUGCCAGCCGGCGUCUGUGUUACGCUUGUGGCAGAUGAAAUGCCAC-CGGCCCCGUC-UACGUUCUUUGGUUGCCUCC-- ---.((((((((((......((((((.((..((((.....))))....)).)))))).....))))))))))........((.((-(((...((..-..))....))))).))....-- ( -38.20, z-score = -1.19, R) >droSec1.super_2 909840 108 + 7591821 ---AAGCCACAGGCAAGUCAACAGACACUUCGGCACCAUUACCCAGCCGGCGUCUGUGUUACGUUUGUGGAAGAUGAAAUGCCAC-CGGCC----C-UACGCUCUUUGGUUACCUCC-- ---.(((((.((((..((..((((((...(((((...........))))).))))))...))((..((((...........))))-..)).----.-...)).)).)))))......-- ( -29.90, z-score = -0.23, R) >droSim1.chr3L 532947 112 + 22553184 ---AAGCCACAGGCAAGUCAACAGACACUUCGACACCAAUUUCCAGCCGGCGUCUGUGUUACGUUUGUGGAAGAUGAAAUGCCAC-CGGCCCUGUC-UACGCUCUUUGGUUACCUCC-- ---.(((((.((((..(((....))).....((((.((.((((((.(..((((.......))))..))))))).))....(((..-.)))..))))-...)).)).)))))......-- ( -28.00, z-score = 0.23, R) >consensus ___AAGCCACAGGCAAGUCAACAGACACCUCGGCACCAAUU_CCAGCCGGCGUCUGUGUUACGUUUGUGGCAGAUGAAAUGCCAC_CGGCCUCGUC_UACGUUCAUUGGUUGCCUCC__ ......((((((((......((((((...(((((...........))))).)))))).....))))))))................................................. (-22.84 = -22.25 + -0.59)
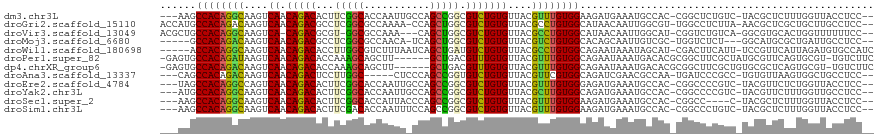
| Location | 890,271 – 890,383 |
|---|---|
| Length | 112 |
| Sequences | 12 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 70.64 |
| Shannon entropy | 0.63498 |
| G+C content | 0.54318 |
| Mean single sequence MFE | -38.28 |
| Consensus MFE | -17.92 |
| Energy contribution | -18.04 |
| Covariance contribution | 0.13 |
| Combinations/Pair | 1.40 |
| Mean z-score | -0.94 |
| Structure conservation index | 0.47 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.38 |
| SVM RNA-class probability | 0.667290 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
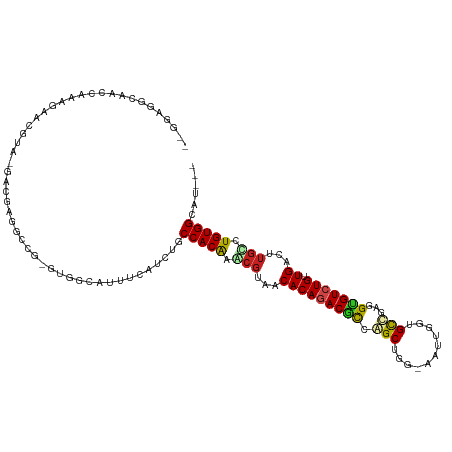
>dm3.chr3L 890271 112 - 24543557 --GGAGGUAACCAAAGAGCGUA-GACAGAGCCG-GUGGCAUUUCAUCUUCCACAAACGUAACACAGACGCCGGCUGGCAAUUGGUGCCGAAGUGUCUGUUGACUUGCCUGUGGCUU--- --((......))..........-....((((((-..((((..(((......((....))...((((((((((((...........))))..)))))))))))..))))..))))))--- ( -33.20, z-score = 0.51, R) >droGri2.scaffold_15110 17215528 114 - 24565398 --GGAGGCAAGCAGCGAGCGUU-UAAGAGGCCA-ACGCCAAUUGUUAUGCCACAGGCGUAACACAGACGCCAGCUGG-UUUUGGCGCCGAGGCGUCUGUUGACUUGUCUGUGGCAUGGU --....((((((.....))...-.....(((..-..)))..))))(((((((((((((....(((((((((...(((-(......)))).))))))))).....))))))))))))).. ( -46.90, z-score = -1.90, R) >droVir3.scaffold_13049 15442492 110 - 25233164 --GGAAAAAACCAGUGCACGCC-UGACAGACCG-AUGCCAAUUGUUAUGCCACAGGCGUAACACAGACGCCAGCUG---UUUGGCGCC-ACGCGUCUG-UGACUUGCCUGUGGCAGCGU --((......)).....((((.-((((((....-.......))))))(((((((((((...(((((((((..(((.---...)))...-..)))))))-))...))))))))))))))) ( -46.40, z-score = -3.02, R) >droMoj3.scaffold_6680 3348614 107 + 24764193 --GGAGGCAAUCAGCGCAUGCC---AGAGACCA-GCGACAAUUGCUGUGCCACAGACGUAACACAGACGCCAGCUGA-UGUUGGCGCCGAGGCGUCUGUUGACUUGUCUGUGGC----- --...((((((...(((.((..---......))-)))...))))))..((((((((((....((((((((((((...-.))))))((....)))))))).....))))))))))----- ( -48.80, z-score = -3.29, R) >droWil1.scaffold_180698 5597282 112 - 11422946 GAUGGCACAUCUAAUGAACGGA-AAUGAAGUCG-AUGCUAUUUAUUCUGCCACAGGCGUAACACAGACAUCAGCUGAUUAAAGACGCCAAGGUGUCUGUUGACUUGCCUGUGGU----- (((((((..((....)).(((.-.......)))-.)))))))......((((((((((....(((((((((.((...........))...))))))))).....))))))))))----- ( -35.90, z-score = -2.41, R) >droPer1.super_82 161585 111 + 267963 GAAGACA-ACGCACUGAACGCAUAGCGAAGCCGCGUGUCAUUUAUUCUGCCACAAACGUAACACAAACGUCAGC------AAGCUGCUUUGGUGUCUGUUGACUUAUCUGUGGCACUC- ...((((-.(((.((...(((...))).))..)))))))........(((((((...((.......))((((((------(..(.((....)))..))))))).....)))))))...- ( -31.80, z-score = -0.76, R) >dp4.chrXR_group6 6491456 111 - 13314419 GAAGACA-ACGCACUGAGCGCACAGCGAAGCCGCGUGUCAUUUAUUCUGCCACAAACGUAACACAAACGUCAGC------AAGCUGCUUUGGUGUCUGUUGACUUGUCUGUGGCACUC- ...((((-.(((.((..((.....))..))..)))))))........(((((((.((((.......))((((((------(..(.((....)))..)))))))..)).)))))))...- ( -35.80, z-score = -0.70, R) >droAna3.scaffold_13337 16266106 107 + 23293914 --GGAGGCAGCCACUUAACACA-GGCGGGAUCA-UUGGCGUUCGAUCUGCCACGAACGUAACACAGACACCGGCUGGGAG-----GCCAAGGAGUCUGUUGACUUGUCUGUGGCUG--- --.....(((((((.......(-((((((.(((-...(((((((........)))))))...((((((.((((((....)-----)))..)).)))))))))))))))))))))))--- ( -42.91, z-score = -2.31, R) >droEre2.scaffold_4784 897336 112 - 25762168 --GGAGGUAACCAGAGAACGUA-GACGGGGCCG-GUGGCAUUUCAUCUCCCACAAACGUAACACAGACGCCGGCUGGCAAUUGGUGCCGAAGUGUCUGUUGACUGGCCUGUGGCUA--- --..(((...((((...((((.-....(((..(-((((....))))).)))....))))...((((((((((((...........))))..))))))))...))))))).......--- ( -34.10, z-score = 1.19, R) >droYak2.chr3L 869544 112 - 24197627 --GGAGGCAACCAAAGAACGUA-GACGGGGCCG-GUGGCAUUUCAUCUGCCACAAGCGUAACACAGACGCCGGCUGGCAAUUGGUGCCGAAGUGUCUGUUGACUUGCCUGUGGCAU--- --((((((.(((......((..-..)).....)-)).)).))))...(((((((.(((....((((((((((((...........))))..)))))))).....))).))))))).--- ( -40.00, z-score = -0.35, R) >droSec1.super_2 909840 108 - 7591821 --GGAGGUAACCAAAGAGCGUA-G----GGCCG-GUGGCAUUUCAUCUUCCACAAACGUAACACAGACGCCGGCUGGGUAAUGGUGCCGAAGUGUCUGUUGACUUGCCUGUGGCUU--- --((......))..........-(----(((((-..((((..(((......((....))...((((((((((((...........))))..)))))))))))..))))..))))))--- ( -32.40, z-score = 0.84, R) >droSim1.chr3L 532947 112 - 22553184 --GGAGGUAACCAAAGAGCGUA-GACAGGGCCG-GUGGCAUUUCAUCUUCCACAAACGUAACACAGACGCCGGCUGGAAAUUGGUGUCGAAGUGUCUGUUGACUUGCCUGUGGCUU--- --........(((.((.(((..-(((((((..(-((((....)))))..)).........((((.((((((((.(....)))))))))...)))))))))....))))).)))...--- ( -31.20, z-score = 0.94, R) >consensus __GGAGGCAACCAAAGAACGUA_GACGAGGCCG_GUGGCAUUUCAUCUGCCACAAACGUAACACAGACGCCAGCUGG_AAUUGGUGCCGAGGUGUCUGUUGACUUGCCUGUGGCAU___ .................................................(((((.(((...(((((((((.(((...........)))...))))))).))...))).)))))...... (-17.92 = -18.04 + 0.13)
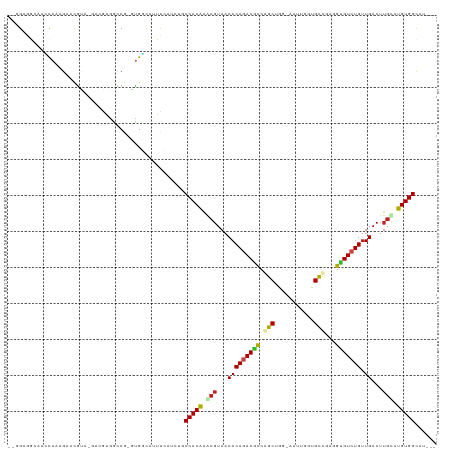
| Location | 890,308 – 890,417 |
|---|---|
| Length | 109 |
| Sequences | 12 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 67.48 |
| Shannon entropy | 0.67493 |
| G+C content | 0.54684 |
| Mean single sequence MFE | -41.93 |
| Consensus MFE | -16.73 |
| Energy contribution | -13.14 |
| Covariance contribution | -3.59 |
| Combinations/Pair | 2.30 |
| Mean z-score | -1.95 |
| Structure conservation index | 0.40 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.99 |
| SVM RNA-class probability | 0.978253 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 890308 109 + 24543557 UGCCAGCCGGCGUCUGUGUUACGUUUGUGGAAGAUGAAAUGCCAC-CGGCUCUGUC-UACGCUCUUUG-GUUACCUC--CGUUUUGGCGGU-AGCU---GCUGAGUGUUGGGGUUGGU- .(((....)))....(((((.(((((.....))))).))))).((-(((((((...-.((((((..((-((((((.(--(.....)).)))-))))---)..)))))).)))))))))- ( -40.60, z-score = -1.58, R) >droGri2.scaffold_15110 17215568 111 + 24565398 -ACCAGCUGGCGUCUGUGUUACGCCUGUGGCAUAACAAUUGGCGU-UGGCCUCUUA-AACGCUCGCUG-CUUGCCUC--CGUUUUGGAUGC-AAGCUUUGCAUAACGUCGAGGCCGAU- -.......((((((..(((((.(((...))).)))))...)))))-)((((((...-.(((...((.(-(((((.((--(.....))).))-))))...))....))).))))))...- ( -43.20, z-score = -2.45, R) >droVir3.scaffold_13049 15442530 110 + 25233164 ---CAGCUGGCGUCUGUGUUACGCCUGUGGCAUAACAAUUGGCAU-CGGUCUGUCA-GGCGUGCACUG-GUUUUUUC--CGUCUUGGACUUGAAUCUUUGCUUAACGUCUAGGUCGAU- ---..((..(....((((((((....))))))))....)..))((-((..((...(-((((((((..(-((((..((--(.....)))...)))))..)))...))))))))..))))- ( -33.90, z-score = -1.29, R) >droMoj3.scaffold_6680 3348649 108 - 24764193 -AUCAGCUGGCGUCUGUGUUACGUCUGUGGCACAGCAAUUGUCGC-UGGUCU--CU-GGCAUGCGCUG-AUUGCCUC--CGUUUUGGACGU-AGUCUGCGCU-GAUGUCGAGGCCAAC- -...(((.((((.(((((((((....)))))))))....))))))-)(((((--(.-(((((((((.(-(((((.((--(.....))).))-)))).)))).-.)))))))))))...- ( -49.70, z-score = -3.95, R) >droWil1.scaffold_180698 5597317 115 + 11422946 AAUCAGCUGAUGUCUGUGUUACGCCUGUGGCAGAAUAAAUAGCAU-CGACUUCAUU-UCCGUUCAUUA-GAUGUGCCAUCGUUUUGAUGGUCAUCCAAUGCUUAACGUUGAGGCCACC- ........(((((...((((..(((...)))..))))....))))-)(.(((((..-..((((((((.-((((.((((((.....)))))))))).))))...)))).))))).)...- ( -30.30, z-score = -1.15, R) >droPer1.super_82 161619 112 - 267963 -GCUUGCUGACGUUUGUGUUACGUUUGUGGCAGAAUAAAUGACACGCGGCUUCGCUAUGCGUUCAGUGCGUUGUCUU--CGUUCUGACGGC-AAUC---GCUGGGCGUUGGAGUCGCGU -(((.((.(((((.......))))).)))))............(((((((((((....((((((((((.((((((.(--(.....)).)))-))))---)))))))))))))))))))) ( -43.80, z-score = -1.70, R) >dp4.chrXR_group6 6491490 112 + 13314419 -GCUUGCUGACGUUUGUGUUACGUUUGUGGCAGAAUAAAUGACACGCGGCUUCGCUGUGCGCUCAGUGCGUUGUCUU--CGUUCUGACGGC-AAUC---GCUGGGCGUUGGAGUCGCGU -(((.((.(((((.......))))).)))))............(((((((((((....((((((((((.((((((.(--(.....)).)))-))))---)))))))))))))))))))) ( -46.30, z-score = -1.92, R) >droAna3.scaffold_13337 16266138 109 - 23293914 UCCCAGCCGGUGUCUGUGUUACGUUCGUGGCAGAUCGAACGCCAA-UGAUCCCGCC-UGUGUUAAGUG-GCUGCCUC--CGUUCUGGCUGC-AACC---AUGCAACACUGGGAUUGUU- ........(((((.((((((((....))))))...)).)))))((-..((((((..-.(((((..(((-(.(((..(--(.....))..))-).))---))..)))))))))))..))- ( -37.60, z-score = -1.25, R) >droEre2.scaffold_4784 897373 109 + 25762168 UGCCAGCCGGCGUCUGUGUUACGUUUGUGGGAGAUGAAAUGCCAC-CGGCCCCGUC-UACGUUCUCUG-GUUACCUC--CGUUUUGGCGGC-AGCU---GCUGAGCGUUGGGGCUGGU- .(((....)))....(((((.(((((.....))))).))))).((-((((((((..-.((((((..((-(((.((.(--(.....)).)).-))))---)..))))))))))))))))- ( -42.70, z-score = -0.51, R) >droYak2.chr3L 869581 109 + 24197627 UGCCAGCCGGCGUCUGUGUUACGCUUGUGGCAGAUGAAAUGCCAC-CGGCCCCGUC-UACGUUCUUUG-GUUGCCUC--CGUUUUGGCGGU-AGCU---GCUGAGCGUUGGGGCUGGU- (((((.(.(((((.......))))).))))))...........((-((((((((..-.((((((..((-((((((.(--(.....)).)))-))))---)..))))))))))))))))- ( -51.70, z-score = -3.07, R) >droSec1.super_2 909877 105 + 7591821 ACCCAGCCGGCGUCUGUGUUACGUUUGUGGAAGAUGAAAUGCCAC-CGGCC----C-UACGCUCUUUG-GUUACCUC--CGUUUUGACGGU-AGCU---GCUGAGUGUUGGGGUUGGU- .....((...(((((...((((....)))).)))))....)).((-(((((----(-(((((((..((-((((((((--......)).)))-))))---)..))))).))))))))))- ( -40.50, z-score = -2.10, R) >droSim1.chr3L 532984 109 + 22553184 UUCCAGCCGGCGUCUGUGUUACGUUUGUGGAAGAUGAAAUGCCAC-CGGCCCUGUC-UACGCUCUUUG-GUUACCUC--CGUUUUGGCGGU-AGCU---GCUGAGUGUUGGGGUUGGU- (((((.(..((((.......))))..))))))...........((-(((((((...-.((((((..((-((((((.(--(.....)).)))-))))---)..)))))).)))))))))- ( -42.90, z-score = -2.38, R) >consensus _GCCAGCCGGCGUCUGUGUUACGUUUGUGGCAGAUGAAAUGCCAC_CGGCCUCGUC_UACGUUCAUUG_GUUGCCUC__CGUUUUGGCGGU_AGCC___GCUGAGCGUUGGGGUCGGU_ ..((.((.(((((.......))))).))))................(((((((.....(((((..........((..........))((((........))))))))).)))))))... (-16.73 = -13.14 + -3.59)

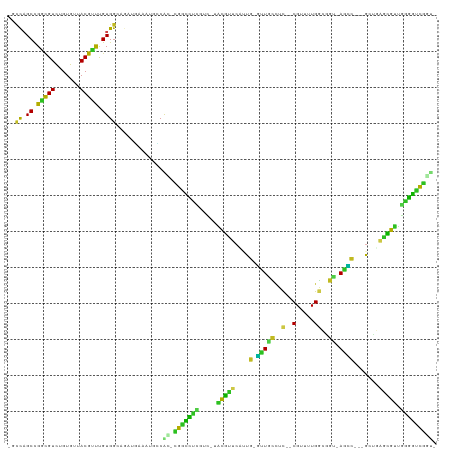
| Location | 890,348 – 890,448 |
|---|---|
| Length | 100 |
| Sequences | 11 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 56.45 |
| Shannon entropy | 0.88061 |
| G+C content | 0.54403 |
| Mean single sequence MFE | -39.63 |
| Consensus MFE | -12.72 |
| Energy contribution | -9.78 |
| Covariance contribution | -2.94 |
| Combinations/Pair | 2.50 |
| Mean z-score | -2.49 |
| Structure conservation index | 0.32 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.34 |
| SVM RNA-class probability | 0.998384 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 890348 100 + 24543557 -GCCACCGGCUCUGUC-UACGCUCUUUG-GU-UACCUCCGUUUUGGCGGUAGCU----GCUGAGUGUUGGGGUUG-GUGCACGUGCUC--CGGUUUUAA---GUAGAAGAUCUC -((.(((((((((...-.((((((..((-((-((((.((.....)).)))))))----)..)))))).)))))))-))))........--.((((((..---....)))))).. ( -37.60, z-score = -2.05, R) >droGri2.scaffold_15110 17215607 103 + 24565398 -GGCGUUGGCCUC-UUAAACGCUCGCUG-CU-UGCCUCCGUUUUGGAUGC-AAGCUUUGCAUAACGUCGAGGCCG-AUGCAAGUGCAUUGUGUAUAUACA-CAUGGAUGA---- -.(((((((((((-....(((...((.(-((-(((.(((.....))).))-))))...))....))).)))))))-))))..((.(((.((((....)))-)))).))..---- ( -42.90, z-score = -3.80, R) >droVir3.scaffold_13049 15442567 102 + 25233164 -GGCAUCGGUCUG-UCAGGCGUGCACUG-GU-UUUUUCCGUCUUGGACUUGAAUCUUUGCUUAACGUCUAGGUCG-AUGCAGGUGCAUGGUGUAUA--CU-CAUGAUUAA---- -.((((((..((.-..(((((((((..(-((-((..(((.....)))...)))))..)))...))))))))..))-)))).....((((((....)--).-)))).....---- ( -32.90, z-score = -2.29, R) >droMoj3.scaffold_6680 3348688 98 - 24764193 -GUCGCUGGUC---UCUGGCAUGCGCUG-AU-UGCCUCCGUUUUGGACGU-AGUCUGCGCU-GAUGUCGAGGCCA-ACGCAAGUGCACGGUGCGUA--CG-CAUGGUUGG---- -...(((((((---((.(((((((((.(-((-(((.(((.....))).))-)))).)))).-.))))))))))))-((....))))((.((((...--.)-))).))...---- ( -44.30, z-score = -2.38, R) >droWil1.scaffold_180698 5597357 111 + 11422946 -AGCAUCGACUUCAUU-UCCGUUCAUUAGAUGUGCCAUCGUUUUGAUGGUCAUCCAAUGCUUAACGUUGAGGCCA-CCGCACGUGUCCAUCGAUAAUAAUGCAUAGACAAUCUC -.((((.....((.((-..((((((((.((((.((((((.....)))))))))).))))...))))..))((.((-(.....))).))...)).....))))............ ( -26.40, z-score = -1.01, R) >droPer1.super_82 161658 106 - 267963 GACACGCGGCUUCGCUAUGCGUUCAGUGCGU-UGUCUUCGUUCUGACGGCAAUC----GCUGGGCGUUGGAGUCGCGUGCACGUGCUCUGCUCUUUCCA---GCAGAGAGUCUU (.((((((((((((....((((((((((.((-((((.((.....)).)))))))----))))))))))))))))))))))((...(((((((......)---)))))).))... ( -50.40, z-score = -3.45, R) >dp4.chrXR_group6 6491529 106 + 13314419 GACACGCGGCUUCGCUGUGCGCUCAGUGCGU-UGUCUUCGUUCUGACGGCAAUC----GCUGGGCGUUGGAGUCGCGUGCACGUGCUCUGCUCUUUCCA---GCAGAGAGUCUU (.((((((((((((....((((((((((.((-((((.((.....)).)))))))----))))))))))))))))))))))((...(((((((......)---)))))).))... ( -52.90, z-score = -3.45, R) >droAna3.scaffold_13337 16266178 97 - 23293914 -GCCAAUGAUCCCGCC-UGUGUUAAGUG-GC-UGCCUCCGUUCUGGCUGCAACC----AUGCAACACUGGGAUUG-UUGCAAGUGCCU--CUCUUU------ACUGAAAGUAUC -((.((..((((((..-.(((((..(((-(.-(((..((.....))..))).))----))..)))))))))))..-))))..((((..--.((...------...))..)))). ( -29.90, z-score = -1.98, R) >droEre2.scaffold_4784 897413 100 + 25762168 -GCCACCGGCCCCGUC-UACGUUCUCUG-GU-UACCUCCGUUUUGGCGGCAGCU----GCUGAGCGUUGGGGCUG-GUGCACGUGCUC--CGAGUUUCA---GCAGAGGAUCUC -((.((((((((((..-.((((((..((-((-(.((.((.....)).)).))))----)..))))))))))))))-))))........--.(((.(((.---.....))).))) ( -40.70, z-score = -0.91, R) >droSec1.super_2 909917 96 + 7591821 -GCCACCGGCCCU-----ACGCUCUUUG-GU-UACCUCCGUUUUGACGGUAGCU----GCUGAGUGUUGGGGUUG-GUGCACGUGCUC--CGAUUUUAA---GUAGAAGAUAUC -((.(((((((((-----((((((..((-((-((((((......)).)))))))----)..))))).))))))))-))))........--.........---............ ( -36.70, z-score = -2.79, R) >droSim1.chr3L 533024 100 + 22553184 -GCCACCGGCCCUGUC-UACGCUCUUUG-GU-UACCUCCGUUUUGGCGGUAGCU----GCUGAGUGUUGGGGUUG-GUGCACGUGCUC--CGAUUUUAA---GUAGAAGAUCUC -((.(((((((((...-.((((((..((-((-((((.((.....)).)))))))----)..)))))).)))))))-))))........--.((((((..---....)))))).. ( -41.20, z-score = -3.27, R) >consensus _GCCACCGGCCCCGUC_UACGUUCACUG_GU_UGCCUCCGUUUUGGCGGUAACU____GCUGAGCGUUGGGGUCG_GUGCACGUGCUC__CGAUUUUAA___GUAGAAGAUCUC ......(((((((.....(((((...........((........))((((........))))))))).)))))))...((....))............................ (-12.72 = -9.78 + -2.94)

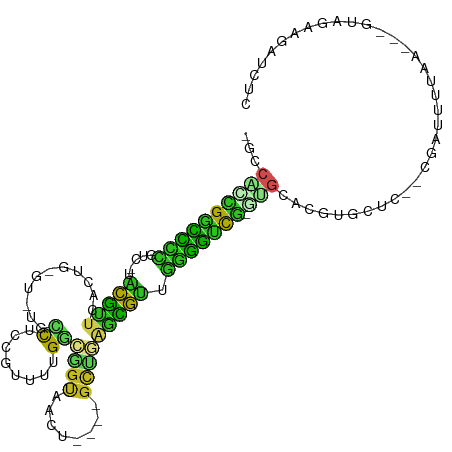
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:54:34 2011